Abstract
Homing, engraftment and proliferation of hematopoietic stem/progenitor cell (HSC/HPCs) are crucial steps required for success of a bone marrow transplant. Observation of these critical events is limited by the opaque nature of bone. Here we demonstrate how individual HSCs engraft in long bones by thinning one side of the tibia for direct and unbiased observation. Intravital imaging enabled detailed visualization of single Sca-1+, c-Kit+, Lineage− (SKL) cell migration to bone marrow niches and subsequent proliferation to reconstitute hematopoiesis. This longitudinal study allowed direct observation of dynamic HSC/HPC activities during engraftment in full color for up to six days in live recipients. Individual SKL cells, but not mature or committed progenitor cells, preferentially homed to a limited number of niches near highly vascularized endosteal regions, and clonally expanded. Engraftment of SKL cells in P-selectin and osteopontin knockout mice showed abnormal homing and expansion of SKL cells. CD150+, CD48− SKL populations initially engrafted in the central marrow region, utilizing only a subset of niches occupied by the parent SKL cells. Our study demonstrates that time-lapse imaging of tibia can be a valuable tool to understand the dynamic characteristics of functional HSC and niche components in various mouse models.
INTRODUCTION
Hematopoietic stem cells (HSCs) are functionally defined by their abilities for clonal proliferation and multi-lineage reconstitution of all blood cells following bone marrow transplantation (BMT). Intravenously infused donor-derived HSCs engraft within the BM niche, expand, and reestablish blood cell production in lethally irradiated recipients. Repopulating HSCs undergo asymmetric cell fate decisions resulting in generation of both new quiescent HSCs – self-renewal - and highly proliferative/differentiating progenitors that eventually restore homeostasis.(1) With extensive studies on hematopoietic cell hierarchy and myeloablative conditioning regimes such as irradiation, the HSC is phenotypically well-defined and tested to the point that a single HSC – isolated in a variety of ways - has been successfully used for long-term reconstitution of the hematopoietic system in mice.(2–6) To assess the reconstitution potential of HSC, the majority of studies examined contribution of donor HSC to peripheral blood of lethally irradiated recipients starting one month post-transplant, with long-term reconstitution usually assessed 4–6 months post-transplant. In contrast, current understanding of early HSC homing and engraftment processes has been mainly derived from immunohistologic analysis of engrafted cells in bone marrow (BM) sections.(7) HSCs are postulated to interact with multiple cell types while migrating to the niche in a dynamic manor. A fixed histologic section provides a single snapshot of individual donor cells – potential HSC- in contact with cells of the recipient marrow microenvironment. No information can be inferred about the transient or stable nature of the observed cellular interactions within the niche. More importantly, histology approaches cannot follow the function of potential HSC over time throughout the dynamic engraftment and repopulation processes. In all established HSC enrichment methods, only a fraction of the sorted cells have been shown to be intrinsic HSC.(8) Standard histology analysis cannot determine whether the observed donor cell is a bona fide HSC that can repopulate BM among the heterogeneous cell population transplanted. Therefore, in vivo imaging is a very attractive tool to longitudinally observe and confirm the intrinsic activities of HSCs, such as engraftment and active proliferation in BM of lethally irradiated recipients(1, 9–14)
The goal of this study was to directly observe the key characteristics associated with HSC function in a living animal during the dynamic repopulation process following BMT. Using time-lapse intravital imaging of tibial long bone as described in Figure 1, we sought to directly visualize the functional abilities of homing, engraftment, clonal expansion and asymmetric cell fates that define HSC activity in lethally irradiated animals. We also wanted to determine if the tibia window technique could distinguish differing engraftment dynamics in transgenic mice with defective HSC niches, or with cell HSC enriched populations with altered engraftment patterns previously assessed via histology. To achieve this goal, we transplanted fluorescently-tagged, HSC enriched Sca-1+, c-Kit+, Lineage− (SKL) cells or other test populations to directly observe their engraftment and repopulation dynamics in individual living recipients for up to six days post transplant.
Figure 1. Time-lapse in vivo imaging of the tibia bone to visualize the engraftment of SKL cells.
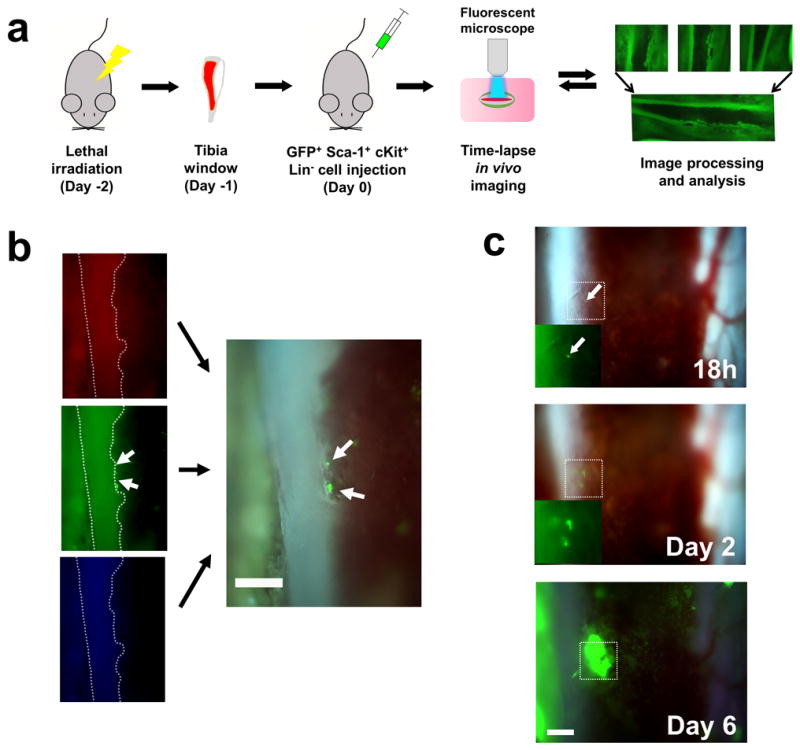
(a) A diagram showing the processes of in vivo tibia imaging. Mice were lethally irradiated 2 days before cell injection. Each image was tiled into a mosaic to create the panoramic view of the tibia marrow. (b) Use of the RGB filter for real-time, true color video recording. Color-separated images from RGB channel demonstrate that individual GFP+ cells could be clearly visualized using 10× magnification (white arrows). Non-fluorescent SKL cells from C57BL6 mice did not generate any signal in the recipients (data not shown). (c) Formation of BM colonies from individual SKL cell in the RGB channel. The fate of a single GFP+ SKL cell was followed over 6 days. All scale bars = 200μm.
MATERIALS AND METHODS
Mice
C57BL6 mice were purchased from Charles River Laboratories (Wilmington, MA). C57BL/6-Tg(UBC-GFP)30Scha/J, Tg(CAG-DsRed*MST)1Nagy/J, FVB/N-TgN(TIE2GFP)287Sato, B6;129S2-Selptm1Hyn (P-selectin knockout), B6.129S6(Cg)-Spp1tm1Blh (OPN knockout) and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NOG) mice were purchased from Jackson Laboratory (Bar Harbor, ME). Tg(CAG-DsRed*MST)1Nagy/J mice were back-crossed with FVB strain for >10 generations at the University of Florida animal facility for BMT into Tie2-GFP recipient mice. Age- and sex-matched animals were randomly grouped for further experiments. All experimental procedures performed on animals were in accordance with the University of Florida’s Institutional Animal Care and Use Committee.
HSC purification and labeling
BM cells from UBC-GFP and DsRed mice for transplantation were age-matched. BM cells were flushed from tibiae and femurs of both legs into PBS supplemented with 1% FBS, 2mM EDTA, 25mM HEPES (MACS buffer). Cells were centrifuged, passed through 20G needle for single cell suspension and filtered through 40μm nylon mesh cell strainer. Cells were treated with Ammonium-Chloride-Potassium (ACK) lysing buffer (0.15M NH4Cl, 1M KHCO3, 0.1mM Na2EDTA, pH 7.2) for 5–10 minutes on ice. SKL populations were isolated as described previously for single-HSC transplants.(3) Antibodies used were mouse Sca-1(clone D7, PE-Cy7), c-Kit (clone 2B8, APC), CD150 (Clone TC15-12 F12.2, Pacific Blue), CD48 (clone HM 48.1), Ter119 (clone Ly-76), Gr-1(clone RB6-8C5), B220 (clone RA3-6B2), CD3 (clone 145 2C11), CD4 (clone L3T4), CD8 (clone 53–6.7). Each population was sorted to >99 % purity via a BD FACSAria II (BD bioscience). For dye retention assays, cells were further stained with DiI dye (Invitrogen; Carlsbad, CA) according to the manufacturers’ protocol and washed with FACS buffer (PBS supplemented with 1.5% FBS) three times before injection.
Irradiation and stem/progenitor cell transplantation
4–5 month old C57BL6 mice were given 950 rads of whole body irradiation using Gamma Cell 40 irradiator (The Atomic Energy of Canada, Ottawa, ON) and subsequently transplanted by retro orbital injection 48 hours following irradiation with 103 to 106 purified cells. Both male and female mice were used as donors and recipients with no overt phenotypic differences observed due to gender. For dye retention assays, 4×103 cells were injected directly through the femoral artery to efficiently deliver the cells into the imaging area. Enrofloxacin (Bayer, Leverkusen, Germany) was added to the drinking water during the first 2 weeks of engraftment to prevent infection.
Tibia window installment
On one day before cell injection (D-1), mice were anesthetized with 90 mg/kg of Ketamine-HCl and 5mg/kg Xylazine-HCl. The hair was removed around the surgical site and the mouse was position on a frame in a supine position under a dissecting microscope (~10-fold magnification). After the mouse skin was disinfected with betadine, a small 5–9 mm incision was made starting from the top of the tibia towards the ankle. The tibia bone was exposed by gently excising the muscle tissue. A sterilized dental drill bit attached to a rotary tool (Dremel, Racine, WI) was used to gently ablate one side of the tibial surface to allow visualization of the marrow through the thinned bone under the dissection microscope.(15, 16) Average of the thickness of the thinned bone of the window was ~40μm, (Supplementary Figure 1). Animals were maintained on analgesia and the window was dressed with antibiotic ointment and a bandage between imaging sessions. As the invasive process of creating the tibia window may alter observable engraftment kinetics, recipients were anesthetized and assessed at 12–24 hours post window installation for minimal inflammatory response and overall clarity of each window. Animals found with window damage or surgical complications during in vivo imaging were not used for subsequent transplant studies. Animals were individually housed and carefully observed for normal movement and eating patterns to ensure minimal distress during the experiment.
In vivo imaging
The initial in vivo imaging sessions were initially performed directly after tibia window installation while the animals were still sedated to confirm transparency of the imaging area. This also established reference position registration hallmarks along each tibia marrow for longitudinal observation. Additional engraftment observation was usually done within 12 hours to 6 days after tibia window installation. Individual observation sessions tended to last from 30 to 45 minutes. Mice were anesthetized with avertin (250 mg/kg, Sigma-Aldrich; St. Louis, MO) prior to in vivo imaging as it increased the survival rate of the repeated in vivo imaging in lethally irradiated recipients. Animals were put on a disinfected and heated stage designed for the microscope use. A few drops of saline were applied on tibia imaging area to prevent drying. Full color images and videos were acquired with 5, 10X magnification air and 20X magnification water lens (Apo, 0.15, 0.40, 0.50 NA respectively) at room temperature using DM5500B microscope (Leica Microsystems, Buffalo Grove, IL), 3 CCD color camera (Hamamatsu C7780; Hamamatsu, Japan) and Volocity 5.5 software (PerkinElmer, Waltham, MA). After each imaging session, the open area was closed with wound clips or surgical suture (Ethicon, Somerville, MA) and plastic bandage was applied around the leg to prevent damage. Some images and 3D images were iteratively restored by deconvolution (confidence limit 100%, Iteration limit 40) using Volocity software. For the whole tibia imaging, 5–8 overlapping individual images of 3.5mm×2.5mm from 5× magnification lens were processed and stitched together using MosaicJ (Biomedical Imaging Group, http://bigwww.epfl.ch/thevenaz/mosaicj/).
Engrafted cell FACS analysis
Animals were sacrificed from 24 hours to day 14 after the cell injection. BM cells were isolated as described above. For animals used in Figure 6, BM cells from the central and the endosteal marrow were separated differentially as described.(17) Both cell populations were treated with ACK buffer on ice for 5–10 min to remove red cells. After the cell number determination, BM cells were re-suspended into FACS buffer and analyzed using BD FACS Canto II (BD bioscience).
Figure 6. Dye retaining cells are located on the endosteal region.
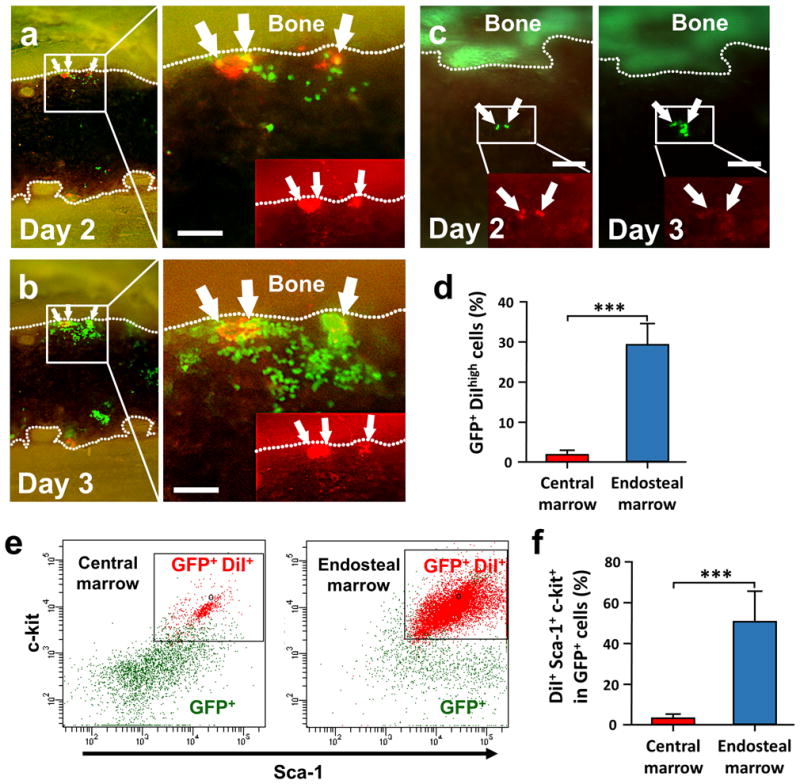
(a and b) Time-lapse in vivo tracking of dye retaining cells to visualize slow cycling cells (n=5). 4×103 GFP+ SKL cells stained with DiI membrane-dye (red) were injected into the femoral artery and monitored for engraftment. GFP+ DiI bright cells were located predominantly near the endosteum (white arrows). (c) The DiI dye that was initially observed in GFP+ SKL cells at the central marrow region at day 2 was rapidly diluted after cell proliferation after 24h. (d–f) FACS analysis of the dye retaining cells from the tibia at day 3 (n=5). Student’s t-test was done for statistical analysis. ***, P<0.001. All scale bars=200μm
Image, video analysis and statistics
Distance and area in the images were calculated using Volocity (Perkin Elmer, Waltham, MA) and ImageJ (NIH, Bethesda, MD) image analysis software. All quantification was performed in a blinded manor to prevent observational bias. All videos were processed and edited with Apple iMovie and Sony Vegas Pro 9.0. For in vivo imaging, each experiment consisted of repeated observations over time of individual animals. Each experimental condition was repeated a minimum of 3 times, normally with 5–10 repetitions to confirm reproducibility. For experiments with statistical analysis, sample numbers were determined to reach statistical significance. Results were analyzed by two-sided Student’s t-test for unpaired groups, or by one-way ANOVA and Tukey’s post-test using Prism 6 (Graphpad, La Jolla, CA). Statistical significance was described in the figure legends and designated as follows: *, P<0.05; **, P<0.01; ***, P<0.001; n.s.= not significant. All values in the data are mean ± SEM.
RESULTS
In vivo imaging of the tibia bone allows direct time-lapse microscopy of fluorescently-tagged HSC engraftment at a single cell level
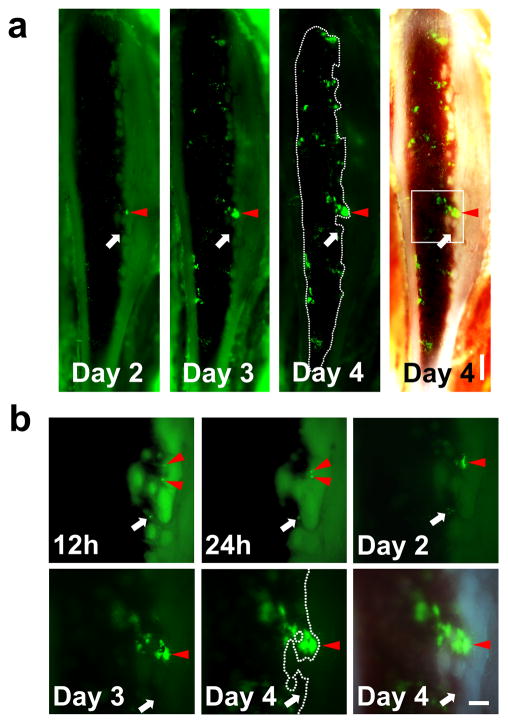
Our initial aim was to selectively visualize key functional stem cell activities within the marrow of an individual living mouse over a period of days following BMT. We chose to use the classic Sca-1+, c-Kit+ Lin− (SKL) BM population isolated from the GFP+ donors. The SKL population is the most widely studied enriched HSC population, but even within SKL studies different laboratories often utilize disparate lineage antibody cocktails, FACS gates etc. We chose to use our SKL isolation protocol that we had previously used for successful single HSC transplants.(3) Our current success rate is 1:5 single cell engrafted recipient animals using this highly HSC enriched SKL population. The SKL population contains both enriched short and long term HSC activity as well as a variety of multipotent progenitors. Engraftment of SKL cells was visualized by surgically thinning one side of the tibia until the bone was sufficiently transparent to allow direct observation of the BM space (Figure 1).(15, 16) Individual frames were tiled to produce mosaic images of the majority of tibia marrow space for unbiased observation (Figures 1 and Supplementary Figure 1). The use of an epifluorescence microscope coupled with triple bandpass RGB filter and 3CCD camera enabled rapid imaging the dynamic process of engraftment in true color and generate real time videos with the single cell level resolution (Figure 1b, c and all Supplementary Videos). Following the BMT, we noticed formation and expansion of engrafted GFP+ colonies mainly along the osteoblastic region of the bone (Figure 2 and Supplementary Video 1). Individual GFP+ SKL cells that homed to and engrafted near the endosteum were routinely observed by 12–48h post-transplant (Figure 2b and Supplementary Figure 2a). Real time video of single GFP+ SKL cells rolling on the endothelium showed that some SKL cells could strongly tether themselves to the endosteal surface (Supplementary Video 2). By 52–60 hours, small but bright GFP+ colonies started to form predominantly along the endosteal region and then rapidly expanded over time to repopulate the marrow space (Figure 2). Time-lapse analysis of individual GFP+ SKL cells also showed that cells engrafted near the endosteal surface could actively expand and dynamically migrate to the surroundings to generate smaller satellite colonies (Figure 3 and Supplementary Video 3). 62.7±8.1% of colonies observed in the tibia window were bordered by the endosteum at day 4 (n=5). We observed that single or a few GFP+ SKL cells that initially engrafted near osterix positive cells (shown to help delineate the potential osteoblastic niche) could proliferate more quickly and form tighter, more populous colonies compared to the few scattered cells and colonies seen in the central marrow region of lethally irradiated recipients (Supplementary Figure 2), which is consistent with previous studies highlighting the importance of osterix in HSC niche formation.(18, 19) SKL enriched cell transplants showed the same overall patterns of engraftment as seen for GFP+ whole bone marrow and/or Lin− control transplants (Supplementary Figure 3 and data not shown).
Figure 2. In vivo imaging of the same mouse over time demonstrates that SKL cells tend to engraft and proliferate near the endosteal region.
(a) Mosaic images of time-lapse in vivo imaging of a tibia window. 3×104 GFP+ SKL cells were injected at Day 0 and in vivo colony formation from SKL cells was monitored in the same animal over time (n=6). Scale bar=500μm (b) Higher magnification of single GFP+ cells engrafted on the endosteal surface developing into a colony (the boxed region at Day 4). Injected SKL cells homed to the marrow and developed into a colony over 4 days (Red arrowheads, the same area marked at the mosaic images). However, not every cell homed to the endosteal surface formed colonies (white arrows). Scale bar=200μm.
Figure 3. Time-lapse imaging of the individual SKL cells showing endosteal engraftment.
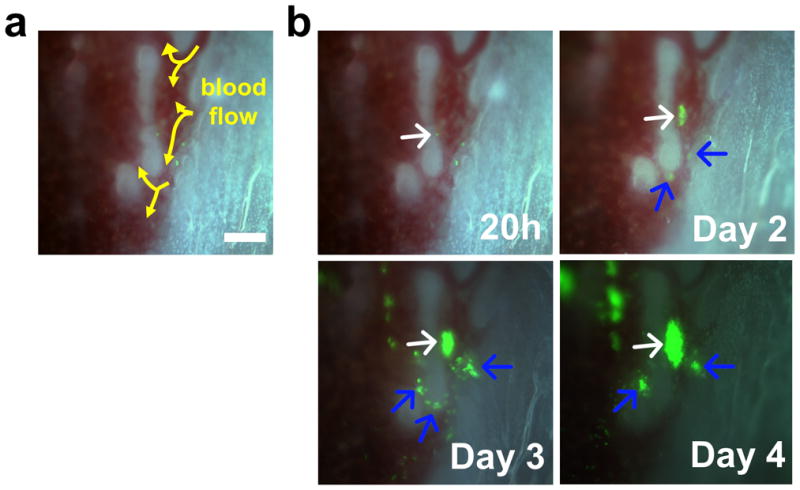
(a) Blood flow (arrows) in the endosteal region. (b) Time lapse imaging of the area showing the engraftment of individual SKL cells. The white arrow indicates the main colony and blue arrows indicate the satellite colonies developed at later time points. Scale bar=100μm.
Early engraftment and proliferation of donor derived cells are stem cell specific
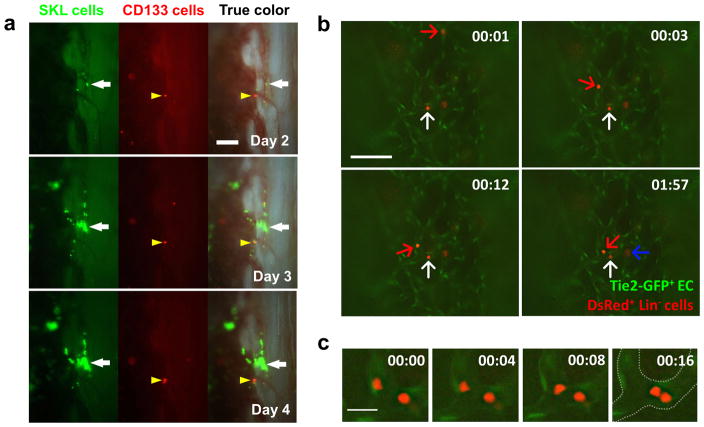
We wanted to directly contrast the homing, engraftment and repopulation patterns of the HSC enriched SKL population versus a known multipotent hematopoietic progenitor population (MPP). We chose CD133+ BM cells from a DsRed transgenic donor as our test MPP population. CD133+ cells were shown to represent a short-term MPP population that can give rise to lymphoid, myeloid and endothelial lineages.(20–22) When both populations were injected, only GFP+ SKL cells were able to actively engraft and proliferate in the BM, preferentially in the endosteal region, which confirmed their homing and repopulation abilities (Figure 4a). A few DsRed+ CD133+ progenitors homed to and engrafted in the marrow, but they failed to proliferate and form colonies. We also transplanted 106 Lin+ GFP+ BM cells in lethally irradiated recipients and followed them for 5 days to understand homing of differentiated cells (Supplementary Figure 4). A few individual Lin+ cells were observed transiently near the central marrow region, but no endosteal engraftment or colony formation was seen. Taken together, the results demonstrate that the SKL population can engraft and rapidly proliferate primarily near the endosteal region, an activity not observed from progenitors or mature cell populations.
Figure 4. Time-lapse and live in vivo imaging to visualize the engraftment process.
(a) Time-lapse in vivo imaging of tibia co-transplanted with 3×104 GFP+ SKL cells (white arrow) and 105 DsRed+ CD133+ hematopoietic progenitor cells (yellow arrowhead, n=3). (b) 5×106 DsRed+ Lin− cells were injected to Tie2-GFP mice to visualize the dynamic interaction of Tie2-GFP+ BM endothelium (EC) and engrafting cells (n=5). Images were simultaneously captured from Video S4 showing rolling/tethering (red arrow) and attaching/extravasating cells (white arrow) that migrate into BM cavity. A cell that is already crossed the endothelial barrier was also observed (blue arrow). Scale bar=200 μm. The observation time is same as appeared in Video S5. (c) Higher magnification images showing cells that tether and scan along the endothelium via millipede-like locomotion. Scale bar = 20μm. Time format = (mm:ss).
Radiation exposure has cytotoxic effects on BM cells that may alter niche usage and location.(23, 24) Therefore, we wanted to examine homing and engraftment of SKL cells under a normal physiological condition. To observe non-ablative engraftment, we injected GFP+ SKL cells into NOD/SCID/IL-2Rγnull (NOG) mice without irradiation.(25) Consistent with ablative transplants, donor SKL cells engrafted predominantly along the endosteal region of NOG mice (Supplementary Figure 5). The colonies that formed in NOG BM were much fewer and tended to be smaller compared to those seen in irradiation chimeras, likely reflecting inherent competition with the native recipient hematopoietic system or lack of non-stem cell facilitator populations during engraftment.(26) Overall, the results suggest that irradiation does not fundamentally alter niche utilization preferences of SKL cells.
Both initial engraftment and proliferation of HSC occur at a limited number of niches with highly vascularized structure
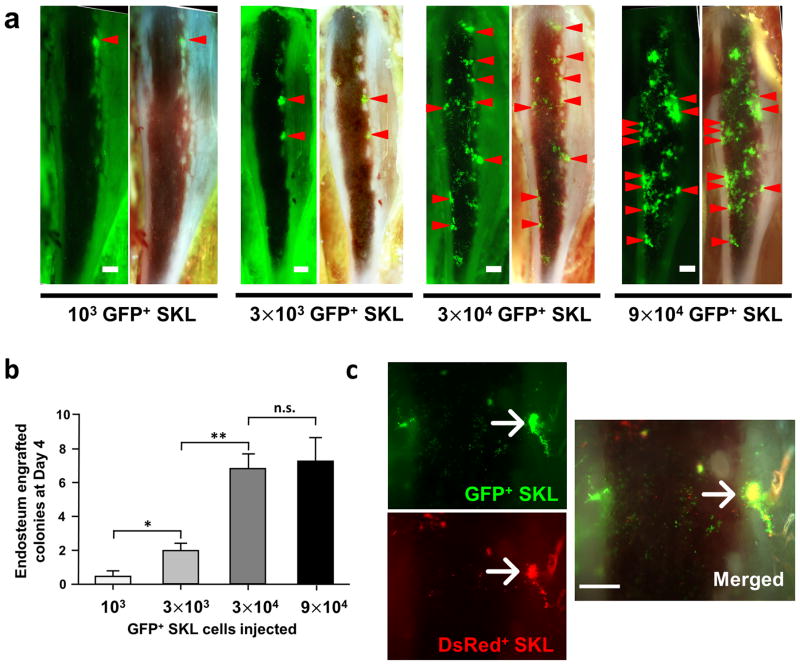
We observed that not every cell that homed to and engrafted at the endosteal surface developed into a colony (white arrows on Figure 2 and Supplementary Video 4). This indicates that many cells within the enriched SKL population may not be functional HSCs, or that otherwise functional HSC may engraft in areas of the marrow that do not support proliferation – perhaps areas that induce quiescence. (1, 27) However, it is also possible that there exists a limited number of distinctive HSC niches within the irradiated marrow capable of supporting functional HSC activity. To address this issue, we performed a series of limiting dilution transplantations with GFP+ SKL cells and demonstrated that when SKL cell numbers were limited, engraftment occurred mostly in the endosteal regions (Figure 5a and Supplementary Video 3). The limited number of engraftment sites was easily saturated, and higher doses of SKL cells did not result in proportional increase of marrow colony number (Figure 5b). In addition, when a saturating dose of both GFP+ SKL cells and DsRed+ SKL cells were injected in the lethally irradiated recipients, we observed that about 1/3 of developing colonies contained both green and red donor cells in the same endosteal regions (8 out of 25 colonies observed, Figure 5c).
Figure 5. SKL cells tend to engraft on the specific niche with the limited number.
(a) Representative images of tibia window with different number of GFP+ SKL cell injection at day 4. Endosteum-engrafted colonies above 3500μm2 were marked with red arrowheads to speculate possible osteoblastic niche locations. (b) The average number of observed colonies in each group was shown in graph (n=5–10). Data analyzed by ANOVA and Tukey’s post test, *, P<0.05; **, P<0.01; n.s.= not significant (c) The same number of SKL cells from GFP and DsRed mice (3×104 cells) were injected into the same recipient to show the niche preference of SKL cells (n=3). Scale bars = 500μm for a, 300μm for c.
HSC homing is initiated by multiple adhesion molecule cascade within BM endothelium and the niche. Selectins are one such adhesion molecule family that mediates tethering and rolling along the endothelium, followed by firm adherence and transendothelial migration.(28) We next aimed to understand the homing process by directly visualizing individual cells real time in tibia BM by injecting 5×106 Lin− donor-cells from DsRed mice to Tie2-GFP recipient mice where the vascular endothelium is GFP+. Among rapidly circulating cells, we could observe a few cells that roll and scan along the Tie2-GFP+ endothelium via millipede-like locomotion (Figure 4c). We also observed cells adhered to the endothelium that are about to exit and other cells that had already migrated to the BM cavity (white and blue arrows in Figure 4b and Supplementary Video 4). The adhesion and extravasation of hematopoietic stem/progenitor cells was only observed at a limited number of locations within the irradiated marrow vasculature.
The HSC niche in irradiated animals is composed of distinct anatomical and functional dimensions to support rapid repopulation of the blood cells.(29, 30) Recent evidence suggests that the vascular/stromal and osteoblastic compartments of the HSC niche may be inseparable.(12, 31) In order to understand the three dimensional interaction between the engrafted and rapidly proliferating SKL cells and the surrounding vasculature, we transplanted DsRed+ SKL cells into Tie2-GFP recipients. Consistent with our previous observation, we found DsRed+ SKL cells engrafted near the endosteum, which became tightly associated with Tie2-GFP+ vascular structures as they developed into BM colonies (Supplementary Figure 6 and Supplementary Video 5). To confirm the in vivo results of HSC engraftment, bones were harvested and stained for MECA32 (Supplementary Figure 6). The GFP+ colonies in the sectioned tibias were also highly vascularized as observed in in vivo imaging. Supplementary Video 4 demonstrates that developing colonies are located in proximity to available blood flow, which would supply oxygen and nutrients to support rapid growth. Taken together, our data provide detailed description of initial homing and engraftment processes that involve recruitment of circulating single HSCs by the BM microvasculature, followed by dynamic trans-marrow migration and expansion processes in supportive niches in living transplant recipients.
DiI dye retaining quiescent cells in proliferating colonies remain in the endosteal area
HSC go through active differentiation in lethally irradiated mice to repopulate the hematopoietic system. HSC were shown to produce asymmetrical cell fates of both relative quiescence (self-renewal) and active proliferation/differentiation (reconstitution) in an ablative transplant environment.(1, 32, 33) One method allowing direct observation of differences in cell proliferation rates among progeny cells is differential cell membrane-dye retention.(34) GFP+ SKL cells were stained with red fluorescent DiI-membrane dye and used for BMT of recipient animals with tibia windows. We initially observed Dil-dye positive cells primarily engrafting in the endosteal area, but occasionally out in the central marrow region (Figures 6a and 6c). However, Dil-dye retaining cells after evidence of extensive GFP+ proliferation/colony formation were only observed near the endosteal region. Dye retaining cells showing little or no proliferation also only persisted near the endosteum by 72 hours post-transplant (Figure 6b). The relative lack of Dil-dye retaining Sca-1+, c-Kit+ cells/progeny outside of the endosteal region beyond 3 days post-transplant was confirmed by FACS analysis (Figure 6d–f) suggesting that the endosteal region selectively maintains slow cycling cells among rapidly dividing daughter cells.
Time-lapse in vivo imaging can be used to understand the important roles of the HSC niche components
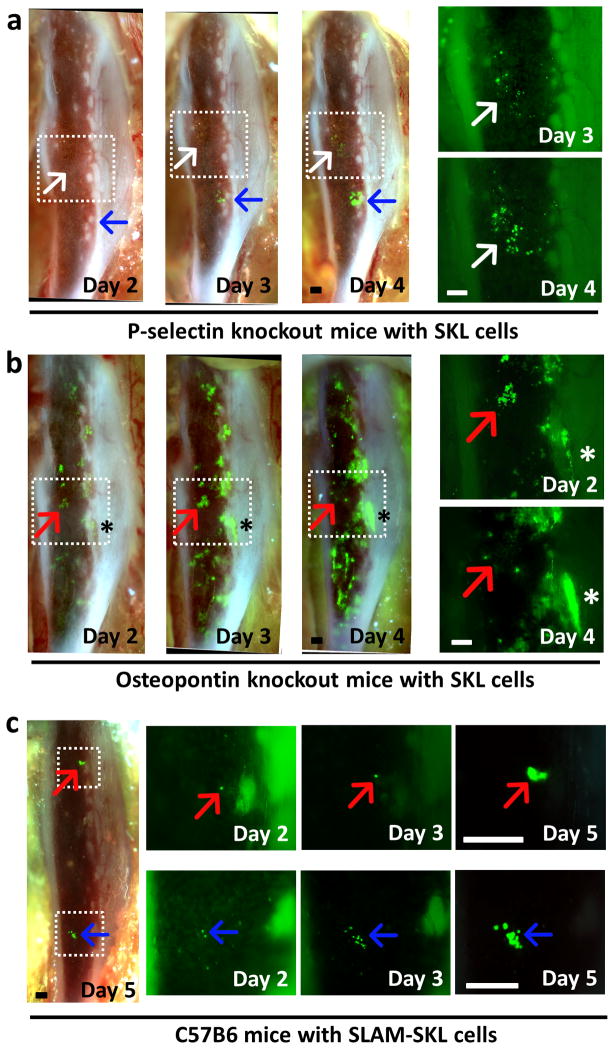
Next, we sought to determine if the tibia window model would facilitate direct observation of altered bone marrow engraftment and niche utilization phenotypes previously documented in the literature. Therefore, we visualized engraftment of GFP+ SKL cells in P-selectin knockout mice to observe HSC engraftment dynamics in the mutant bone marrow microenvironment. P-selectin is one of the major BM endothelial adhesion molecules involved in the initial rolling and extravasation steps on BMT. P-selectin null marrow would be predicted to have negative effects on WT-donor cell engraftment.(35) Indeed, we had to increase the injected cell number to 6×104 SKL cells to ensure survival in these mice as engraftment was very poor. Time-lapse imaging showed that there were small colonies developed in the central region of the P-selectin null marrow, showing that endosteal engraftment is severely compromised by the mutation (Figure 7a). While majority of the engrafted cells were scattered in the central marrow region, a few single SKL cells did engraft near the endosteum at 24–48h (Supplementary Figure 7). These few cells formed clusters at later time points (blue arrows in Figure 7a), and eventually developed into much bigger colonies than others observed in the central marrow regions (white arrows in Figure 7a). Large colony formation was significantly delayed in P-selectin null recipients compared to normal.
Figure 7. SKL cell engraftment in abnormal HSC niches.
(a) Time-lapse in vivo imaging of SKL cell engraftment in P-selectin knockout mice (n=5). Cells engrafted in the central marrow area engrafted and proliferated much slower (white arrow) than a cell that homed to the endosteal region (blue arrow). Higher magnification pictures of the boxed area on the right side show aberrant engraftment in the central marrow region (white arrow). 6×104 GFP+ SKL cells were injected at Day 0 after lethal irradiation. (b) In vivo imaging of SKL cell engraftment in OPN knockout mice showing rapid proliferation of SKL cells (n=4). Higher magnification pictures of the boxed area on the right side show clusters of SKL cells at the central marrow region disappearing at day 3 (red arrow). SKL cells in the endosteal regions showed rapid proliferation (asterisk). 3×104 GFP+ SKL cells were injected at Day 0 after lethal irradiation. (c) 3×103 GFP+ SLAM-SKL cells were injected at Day 0. Engraftment and proliferation of individual SLAM-SKL cells (red/blue arrows) were observed till Day 5.
Osteopontin (OPN) is a key component of the HSC niche that is secreted by osteoblasts. Studies have shown that it acts as a potent constraint on HSC proliferation as OPN-null microenvironment leads to increased HSC proliferation.(36, 37) We also observed rapid proliferation of GFP+ WT-SKL cells in these OPN-null recipient marrows in vivo. Rapidly proliferating colonies occurred mainly near the endosteal regions (Figure 7b). Rapidly proliferating colonies were also initially formed in the central marrow region of these mice, however, many of them dissipated quickly by day 4 (red arrows in Figure 7b). While the observed GFP+ SKL cells were mainly located near the endosteum in C57BL6 mice at 18h after cell injection, the same population in P-selectin and OPN knockout mice was randomly distributed, showing that disruptions of vascular or endosteal HSC niche components result in aberrant homing and engraftment of SKL cells (Supplementary Figure 8).
The addition of signaling lymphocyte activation molecule (SLAM) markers CD150 and CD48 to the SKL enrichment scheme yields a highly purified myeloid biased HSC population that is reported to preferentially utilize the central vascular niche of bone marrow after transplant (4, 38). We isolated GFP+ CD150+, CD48− (SLAM)-SKL cells for in vivo imaging via a tibia window. The SLAM-SKL population showed a contrasting engraftment pattern to the parent SKL population (Figure 7c). While SKL cells predominantly expanded quickly within the endosteal niche, SLAM-SKL cells formed smaller colonies mostly in the central marrow or the perivascular/stromal niche during the observation period. Both populations provided subsequent long-term radioprotection to their recipients demonstrating HSC function (data not shown). The two engrafted HSC populations have distinct patterns of predominant niche utilization, with the SLAM-SKL population utilizing a subset of niches used by the parent SKL population and forming colonies within the marrow at a slower initial rate.
DISCUSSION
In this study, we sought to create a straightforward in vivo imaging model that allowed direct observation of different populations of transplanted cells, with resolution at the individual cell level. Using the tibia window method, we were able to visualize engraftment of SKL cells across the majority of tibia marrow space for unbiased observation. The use of epifluorescence microscopy coupled with a triple bandpass RGB fluorescent filter cube and a full color 3CCD camera made it possible to simultaneously capture fluorescent micrographs of multiple cell types at a full video frame rate. This simple combination also enabled us to monitor the dynamic HSC homing, engraftment and proliferation processes relatively quickly for a given imaging session. The tibia window also facilitated repeated observations over the span of up to a week in individual animals. The functional observation period for a given window was dictated by the speed of bone healing, which made the imaging area opaque. In most cases, observation of the critical engraftment process was possible until the marrow became so reconstituted with donor cells that the overall level of fluorescence obscured the view of cellular activity – usually 4–6 days after BMT.
The HSC is defined by its functional ability to maintain hematopoiesis. The majority of studies of HSC biology have utilized retrospective analyses such as flow cytometry and histologic techniques, i.e. immunohistochemistry. Although FACS can provide serial analysis of peripheral blood composition to assess multi-lineage reconstitution, it does not provide spatial information for engrafted HSC within the marrow. In contrast, immunostaining provides a single snap shot of one instant in time per individual analyzed. Examination of many individual snapshots revealed repeated interactions between donor cells, often inferred to be HSC, with various niche components such as osteoblasts and endothelial/stromal cells. These frequent associations have formed the scientific basis for the concept of osteoblastic and perivascular/stromal niches, which are thought to support HSC activity within the marrow.(11, 12) Although repeated static observations are very suggestive evidence that recipient niche cells and adjacent donor HSC are interacting, it cannot provide a direct proof that any observed cell subsequently exhibits HSC activity. In this regard, directly imaging how functional HSC in a heterogeneous population engraft and repopulate BM of live animals can be a powerful tool for comprehensive analysis of dynamic stem cell activity.(10, 13)
Our major goal was to directly visualize key characteristics associated with HSC function during BMT by following HSC engraftment and expansion processes in the same animal over time. For this purpose, we chose the classic SKL population for initial study. SKL cells are highly enriched for HSC activity yet still contain a spectrum of cells with varying engraftment potentials. Previous studies have shown that as few as 100 SKL cells were sufficient to rescue lethally irradiated mice, demonstrating that the population is highly enriched for ST- and LT-HSC required to rescue the recipients.(8, 39–41) In addition, using single HSC transplant from SKL population, we and others successfully provided proof that SKL cells contain clonal HSC with long-term multi-lineage repopulating potential.(2, 3, 42) We speculated that time-lapse in vivo imaging could demonstrate key functional HSC activities required to repopulate the BM and peripheral blood following transplant in order to reestablish homeostasis in irradiated recipients. We observed in vivo the fate of engrafted individual SKL cells over time for their ability to exhibit the following key classic hallmarks associated with HSC function.
First, using tibia windows we documented donor SKL populations homing to marrow via the circulation where they extravasate into the marrow space (Figure 4, Supplementary Videos 2 and 4). Donor SKL engraft within the tibia long bone with a strong preference for a limited number of niches for both extravasation and engraftment within irradiated recipients (Figures 1–5, Supplementary Videos 1 and 3). The SKL engraftment phenotype was indistinguishable from that observed with either whole bone marrow or mildly enriched Lin− donor populations (Supplementary Figure 3), suggesting that no major stem/progenitor activities were excluded by SKL enrichment. In contrast, Lin+ transplants failed to home to or engraft the marrow to any appreciable degree (Supplemental Figure 2). CD133+ MPP did occasionally engraft, but failed to significantly proliferate within the marrow. These data suggest that the SKL population contains the majority of homing/engraftment/expansion active cell types, or at least niche utilization phenotypes contained in whole marrow. Furthermore, significant marrow colony formation was only observed when populations known to contain HSC activity were transplanted. Thus, the ability to produce colony forming unit-BM (CFU-BM) during marrow repopulation likely represents HSC activity. However, the observable timeframe of the tibia window cannot distinguish short versus long-term HSC populations. Additional test populations containing known CFU and MPP activity without HSC function would be required to further support the CFU-BM as a measure of HSC activity. When comparing population studies, it is always possible that minor populations capable of engraftment and expansion excluded by the SKL enrichment process are not appreciated, especially if their numbers are low and their niche utilization phenotypes duplicate activities contained in the SKL population.
The predominant engraftment site observed with SKL, WBM or Lin− populations was located near the endosteum adjacent to Tie2+ vascular structures, with limited engraftment observed in the central marrow regions (Figure 4 and Supplementary Video 5). Occasional double occupancy in the niche, limiting-dilution and mixed green/red donor SKL experiments suggest that the number of engraftment sites located within the irradiated marrow is limited (Figure 2b and Figure 5). Non-ablative transplants into NOG recipients showed a similar overall pattern of engraftment observed in irradiated transplants, although colony sizes were reduced. This supports the theory that HSC niches are restricted during normal homeostasis as well.
HSC undergo clonal expansion to repopulate both the HSC pool and peripheral blood following myeloablative transplant. The tibia window allowed direct observation of single cell engraftment within the marrow. (Figures 2, 3 and Supplementary Videos 1) The balance between proliferation and quiescence is critical for maintaining the appropriate number of HSCs.(43) Such asymmetric HSC fates were demonstrated by differential membrane dye retention within rapidly expanding GFP marrow colonies. The more quiescent cells were restricted to the endosteal region of the marrow with dye retaining cells being rapidly lost from the central marrow region (Figure 6). Our results suggest that there exist a limited number of specialized endosteal zones, possibly with a distinct blood vessel type for HSC maintenance, that play a critical role in preserving slow cycling stem cells among the rapidly proliferating daughter cells.(44) Although recent studies have highlighted the important roles of BM endothelial cells to HSCs, stem cell activities are still found near the endosteal region of both normal and myeloablated conditions,(17, 44–48) which is in agreement to our observation. As suggested by several reviews, osteoblastic and perivascular/stromal niches may not be mutually exclusive to each other.(49, 50) But rather, they are likely to be physically and functionally involved together to facilitate engraftment of HSC. For instance, metaphyseal spongiosa in femur, where osteoblasts and endothelial/stromal cells are highly mixed, is active HSC engraftment sites. Although our method cannot fully visualize metaphyseal tibia, we also observed many colonies formed on the trabecules of woven tibia bone which supports this idea (Figure 3, Supplementary Video 1 and 3).
The initial phases of homing, engraftment and marrow repopulation are very dynamic and variable processes. The utility of the tibia window model for assessing alterations to early bone marrow engraftment dynamics was demonstrated by the observed altered engraftment patterns in the abnormal HSC niches of P-selectin knockout (deleterious for homing) and OPN knockout (enhanced for proliferation) mice (Figure 7). The tibia window also allows direct observation of altered niche utilization among HSC populations as shown by engraftment of SLAM-SKL cells, which are highly enriched for a myeloid biased LT-HSC (Figure 7c).(38) The SLAM-SKL produced far fewer colonies located away from endosteum proving that the predominant HSC engraftment/repopulation activity of the parent SKL population along the endosteum is not due to HSCs from the SLAM-SKL subpopulation. This indicates each HSC population has different engraftment/proliferation kinetics and further experiments will be required to elucidate cellular dynamics of each population.
This study has several limitations. First, we used heterogeneous SKL cells to selectively observe BM reconstituting HSCs in live animals over time, but this cannot clearly distinguish reconstitution activities among more primitive progenitors, ST- and LT-HSCs. Multi-color reporter mice will be useful donors for this purpose.(51) Second, single cell resolution in an actively proliferating colony is lost within 2–3 days, making long-term observation of the individual HSC/progeny activities difficult. High resolution microscopy will be necessary to directly access cell division at the cellular level. Third, although we had a similar observations in non-invasive MRI HSC homing study(14), HSC engraftment in invasive tibia imaging process may be biased due to bone damage or inflammation. Any potential recipient with window damage or surgical complications was excluded from the study to minimize this possibility.
In vivo imaging of tibia indicates multiple known functional HSC-enriched populations such as SKL cells and SLAM-SKL cells can have different engraftment kinetics and niche utilization in the initial phases of BMT. It should be noted that HSC from antibody mediated selection methods may not be truly representative of the totality of long-term repopulating cells as surface markers of HSC continually change. As a result, these cells may be biased or discarded with the purification processes.(38, 54) Future studies will use the tibia window model to investigate in vivo repopulation dynamics of a variety of previously defined hematopoietic and leukemic stem cells to define their similarities and differences. For instance, HSCs purified via Hoechst/Rhodamine dye retention may have distinct cell cycle and engraftment kinetics.(26, 52, 53) In addition, we have used transgenic mice such as Tie2-GFP, P-selectin and OPN knock-out mice or angiotensin II infusion to demonstrate the feasibility of detailed engraftment observation in altered HSC niche environments.(55) Further studies with the time-lapse in vivo imaging of tibia will unravel the dynamic interaction between the functional HSC and the BM niche components, which will lead to better understanding of the hematopoietic malignancies and BMT.
Supplementary Material
Acknowledgments
Authors thank M. K. Raizada, G. P. Marshall, S.Y. Kim and N. Benson for their discussion and support.
Footnotes
CONFLICT OF INTEREST
Authors declare no conflict of interest.
References
- 1.Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol. 2006 Feb;6(2):93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- 2.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996 Jul 12;273(5272):242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 3.Grant MB, May WS, Caballero S, Brown GA, Guthrie SM, Mames RN, et al. Adult hematopoietic stem cells provide functional hemangioblast activity during retinal neovascularization. Nat Med. 2002 Jun;8(6):607–612. doi: 10.1038/nm0602-607. [DOI] [PubMed] [Google Scholar]
- 4.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005 Jul 1;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 5.Wagers AJ, Sherwood RI, Christensen JL, Weissman IL. Little evidence for developmental plasticity of adult hematopoietic stem cells. Science. 2002 Sep 27;297(5590):2256–2259. doi: 10.1126/science.1074807. [DOI] [PubMed] [Google Scholar]
- 6.Weksberg DC, Chambers SM, Boles NC, Goodell MA. CD150-side population cells represent a functionally distinct population of long-term hematopoietic stem cells. Blood. 2008 Feb 15;111(4):2444–2451. doi: 10.1182/blood-2007-09-115006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purton LE, Scadden DT. StemBook [Internet] Cambridge (MA): Harvard Stem Cell Institute; 2008. The hematopoietic stem cell niche. [PubMed] [Google Scholar]
- 8.Challen GA, Boles N, Lin KK, Goodell MA. Mouse hematopoietic stem cell identification and analysis. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2009 Jan;75(1):14–24. doi: 10.1002/cyto.a.20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiel MJ, Morrison SJ. Uncertainty in the niches that maintain haematopoietic stem cells. Nat Rev Immunol. 2008 Apr;8(4):290–301. doi: 10.1038/nri2279. [DOI] [PubMed] [Google Scholar]
- 10.Lo Celso C, Wu JW, Lin CP. In vivo imaging of hematopoietic stem cells and their microenvironment. J Biophotonics. 2009 Nov;2(11):619–631. doi: 10.1002/jbio.200910072. [DOI] [PubMed] [Google Scholar]
- 11.Ugarte F, Forsberg EC. Haematopoietic stem cell niches: new insights inspire new questions. The EMBO journal. 2013 Sep 10;32(19):2535–2547. doi: 10.1038/emboj.2013.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014 Jan 16;505(7483):327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kokkaliaris KD, Loeffler D, Schroeder T. Advances in tracking hematopoiesis at the single-cell level. Curr Opin Hematol. 2012 Jul;19(4):243–249. doi: 10.1097/MOH.0b013e32835421de. [DOI] [PubMed] [Google Scholar]
- 14.Bengtsson NE, Kim S, Lin L, Walter GA, Scott EW. Ultra-high-field MRI real-time imaging of HSC engraftment of the bone marrow niche. Leukemia. 2011 Aug;25(8):1223–1231. doi: 10.1038/leu.2011.72. [DOI] [PubMed] [Google Scholar]
- 15.Kohler A, Schmithorst V, Filippi MD, Ryan MA, Daria D, Gunzer M, et al. Altered cellular dynamics and endosteal location of aged early hematopoietic progenitor cells revealed by time-lapse intravital imaging in long bones. Blood. 2009 Jul 9;114(2):290–298. doi: 10.1182/blood-2008-12-195644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujisaki J, Wu J, Carlson AL, Silberstein L, Putheti P, Larocca R, et al. In vivo imaging of Treg cells providing immune privilege to the haematopoietic stem-cell niche. Nature. 2011 Jun 9;474(7350):216–219. doi: 10.1038/nature10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haylock DN, Williams B, Johnston HM, Liu MC, Rutherford KE, Whitty GA, et al. Hemopoietic stem cells with higher hemopoietic potential reside at the bone marrow endosteum. Stem Cells. 2007 Apr;25(4):1062–1069. doi: 10.1634/stemcells.2006-0528. [DOI] [PubMed] [Google Scholar]
- 18.Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013 Mar 14;495(7440):227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Y, Yin T, Wiegraebe W, He XC, Miller D, Stark D, et al. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature. 2009 Jan 1;457(7225):97–101. doi: 10.1038/nature07639. [DOI] [PubMed] [Google Scholar]
- 20.Butler JM, Guthrie SM, Koc M, Afzal A, Caballero S, Brooks HL, et al. SDF-1 is both necessary and sufficient to promote proliferative retinopathy. J Clin Invest. 2005 Jan;115(1):86–93. doi: 10.1172/JCI22869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris JR, Fisher R, Jorgensen M, Kaushal S, Scott EW. CD133 progenitor cells from the bone marrow contribute to retinal pigment epithelium repair. Stem Cells. 2009 Feb;27(2):457–466. doi: 10.1634/stemcells.2008-0836. [DOI] [PubMed] [Google Scholar]
- 22.Arndt K, Grinenko T, Mende N, Reichert D, Portz M, Ripich T, et al. CD133 is a modifier of hematopoietic progenitor frequencies but is dispensable for the maintenance of mouse hematopoietic stem cells. Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):5582–5587. doi: 10.1073/pnas.1215438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominici M, Rasini V, Bussolari R, Chen X, Hofmann TJ, Spano C, et al. Restoration and reversible expansion of the osteoblastic hematopoietic stem cell niche after marrow radioablation. Blood. 2009 Sep 10;114(11):2333–2343. doi: 10.1182/blood-2008-10-183459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slayton WB, Li XM, Butler J, Guthrie SM, Jorgensen ML, Wingard JR, et al. The role of the donor in the repair of the marrow vascular niche following hematopoietic stem cell transplant. Stem Cells. 2007 Nov;25(11):2945–2955. doi: 10.1634/stemcells.2007-0158. [DOI] [PubMed] [Google Scholar]
- 25.Waskow C, Madan V, Bartels S, Costa C, Blasig R, Rodewald HR. Hematopoietic stem cell transplantation without irradiation. Nat Methods. 2009 Apr;6(4):267–269. doi: 10.1038/nmeth.1309. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson SK, Dooner MS, Tiarks CY, Weier HU, Quesenberry PJ. Potential and distribution of transplanted hematopoietic stem cells in a nonablated mouse model. Blood. 1997 Jun 1;89(11):4013–4020. [PubMed] [Google Scholar]
- 27.Yin T, Li L. The stem cell niches in bone. J Clin Invest. 2006 May;116(5):1195–1201. doi: 10.1172/JCI28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsson SK, Simmons PJ, Bertoncello I. Hemopoietic stem cell engraftment. Exp Hematol. 2006 Feb;34(2):123–129. doi: 10.1016/j.exphem.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Srour EF, Jetmore A, Wolber FM, Plett PA, Abonour R, Yoder MC, et al. Homing, cell cycle kinetics and fate of transplanted hematopoietic stem cells. Leukemia. 2001 Nov;15(11):1681–1684. doi: 10.1038/sj.leu.2402256. [DOI] [PubMed] [Google Scholar]
- 30.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006 Jun 29;441(7097):1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 31.Krause DS, Scadden DT, Preffer FI. The hematopoietic stem cell niche--home for friend and foe? Cytometry Part B, Clinical cytometry. 2013 Jan-Feb;84(1):7–20. doi: 10.1002/cyto.b.21066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003 Oct 23;425(6960):836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 33.Wu M, Kwon HY, Rattis F, Blum J, Zhao C, Ashkenazi R, et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007 Nov;1(5):541–554. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sipkins DA, Wei X, Wu JW, Runnels JM, Cote D, Means TK, et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005 Jun 16;435(7044):969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazo IB, Gutierrez-Ramos JC, Frenette PS, Hynes RO, Wagner DD, von Andrian UH. Hematopoietic progenitor cell rolling in bone marrow microvessels: parallel contributions by endothelial selectins and vascular cell adhesion molecule 1. J Exp Med. 1998 Aug 3;188(3):465–474. doi: 10.1084/jem.188.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilsson SK, Johnston HM, Whitty GA, Williams B, Webb RJ, Denhardt DT, et al. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood. 2005 Aug 15;106(4):1232–1239. doi: 10.1182/blood-2004-11-4422. [DOI] [PubMed] [Google Scholar]
- 37.Stier S, Ko Y, Forkert R, Lutz C, Neuhaus T, Grunewald E, et al. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med. 2005 Jun 6;201(11):1781–1791. doi: 10.1084/jem.20041992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010 Jun 7;207(6):1173–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikuta K, Weissman IL. Evidence that hematopoietic stem cells express mouse c-kit but do not depend on steel factor for their generation. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1502–1506. doi: 10.1073/pnas.89.4.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li CL, Johnson GR. Murine hematopoietic stem and progenitor cells: I. Enrichment and biologic characterization. Blood. 1995 Mar 15;85(6):1472–1479. [PubMed] [Google Scholar]
- 41.Osawa M, Nakamura K, Nishi N, Takahasi N, Tokuomoto Y, Inoue H, et al. In vivo self-renewal of c-Kit+ Sca-1+ Lin(low/−) hemopoietic stem cells. J Immunol. 1996 May 1;156(9):3207–3214. [PubMed] [Google Scholar]
- 42.Ema H, Morita Y, Yamazaki S, Matsubara A, Seita J, Tadokoro Y, et al. Adult mouse hematopoietic stem cells: purification and single-cell assays. Nat Protoc. 2006;1(6):2979–2987. doi: 10.1038/nprot.2006.447. [DOI] [PubMed] [Google Scholar]
- 43.Trumpp A, Essers M, Wilson A. Awakening dormant haematopoietic stem cells. Nat Rev Immunol. 2010 Mar;10(3):201–209. doi: 10.1038/nri2726. [DOI] [PubMed] [Google Scholar]
- 44.Itkin T, Gur-Cohen S, Spencer JA, Schajnovitz A, Ramasamy SK, Kusumbe AP, et al. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature. 2016 Apr 21;532(7599):323–328. doi: 10.1038/nature17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Acar M, Kocherlakota KS, Murphy MM, Peyer JG, Oguro H, Inra CN, et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 2015 Oct 1;526(7571):126–130. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013 Oct 31;502(7473):637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellis SL, Grassinger J, Jones A, Borg J, Camenisch T, Haylock D, et al. The relationship between bone, hemopoietic stem cells, and vasculature. Blood. 2011 Aug 11;118(6):1516–1524. doi: 10.1182/blood-2010-08-303800. [DOI] [PubMed] [Google Scholar]
- 48.Nombela-Arrieta C, Pivarnik G, Winkel B, Canty KJ, Harley B, Mahoney JE, et al. Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat Cell Biol. 2013 May;15(5):533–543. doi: 10.1038/ncb2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levesque JP, Helwani FM, Winkler IG. The endosteal ‘osteoblastic’ niche and its role in hematopoietic stem cell homing and mobilization. Leukemia. 2010 Dec;24(12):1979–1992. doi: 10.1038/leu.2010.214. [DOI] [PubMed] [Google Scholar]
- 50.Boulais PE, Frenette PS. Making sense of hematopoietic stem cell niches. Blood. Apr 23;125(17):2621–2629. doi: 10.1182/blood-2014-09-570192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, Benedito R, Bixel MG, Zeuschner D, Stehling M, Savendahl L, et al. Identification of a clonally expanding haematopoietic compartment in bone marrow. The EMBO journal. 2013 Jan 23;32(2):219–230. doi: 10.1038/emboj.2012.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldberg LR, Dooner MS, Johnson KW, Papa EF, Pereira MG, Del Tatto M, et al. The murine long-term multi-lineage renewal marrow stem cell is a cycling cell. Leukemia. 2014 Apr;28(4):813–822. doi: 10.1038/leu.2013.252. [DOI] [PubMed] [Google Scholar]
- 53.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996 Apr 1;183(4):1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quesenberry PJ, Goldberg LR, Dooner MS. Concise reviews: A stem cell apostasy: a tale of four H words. Stem Cells. 2015 Jan;33(1):15–20. doi: 10.1002/stem.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim S, Zingler M, Harrison JK, Scott EW, Cogle CR, Luo D, et al. Angiotensin II Regulation of Proliferation, Differentiation, and Engraftment of Hematopoietic Stem Cells. Hypertension. 2016 Mar;67(3):574–584. doi: 10.1161/HYPERTENSIONAHA.115.06474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.