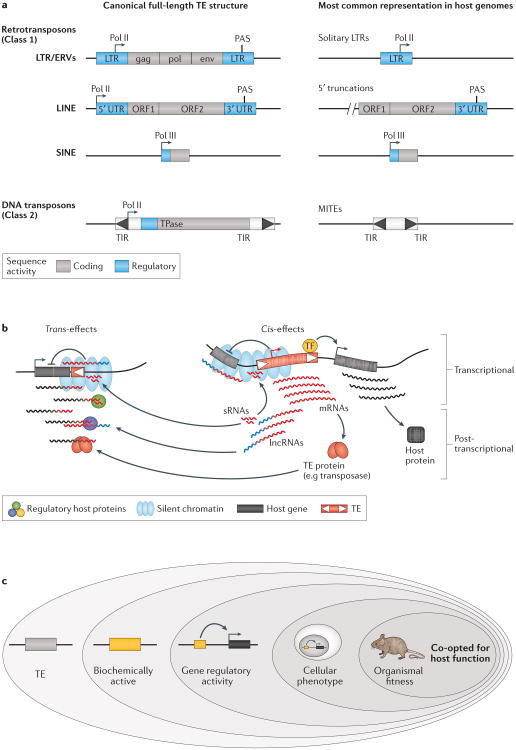
Figure 1. Origins of TE regulatory activities and how they may impact host genes.
a | Schematic of major transposable element (TE) classes and their typical genetic organization. The left panel depicts the idealized full-length version of each TE type. Most TEs harbour regulatory sequences that function to promote their own transcription and regulation, such as promoters for RNA polymerase II (Pol II) or RNA polymerase III (Pol III), and polyadenylation signals (PAS). The right panel shows the structures of each type of TEs as they most commonly occur in the genome, which can differ substantially based on the TE (see the main text for details). b | Diagram depicting different types of regulatory activities exerted by TEs. These include effects mediated by cis-regulatory DNA and RNA elements (right panel) as well as trans effects mediated by TE-produced non-coding RNAs and proteins (left panel). c | Hierarchy of evidence to consider when determining if a TE has been co-opted for host functions. Many TEs exhibit biochemical hallmarks of regulatory activity based on genome-wide assays. However, additional evidence is required to determine which of these TEs alter the regulation of host genes and affect organismal phenotype and fitness. Abbreviations: LTRs: long terminal repeats; ERVs: endogenous retrovirus; MITEs: miniature inverted-repeat transposable elements; sRNAs: small RNAs; TIR: terminal inverted repeats.