Figure 1.
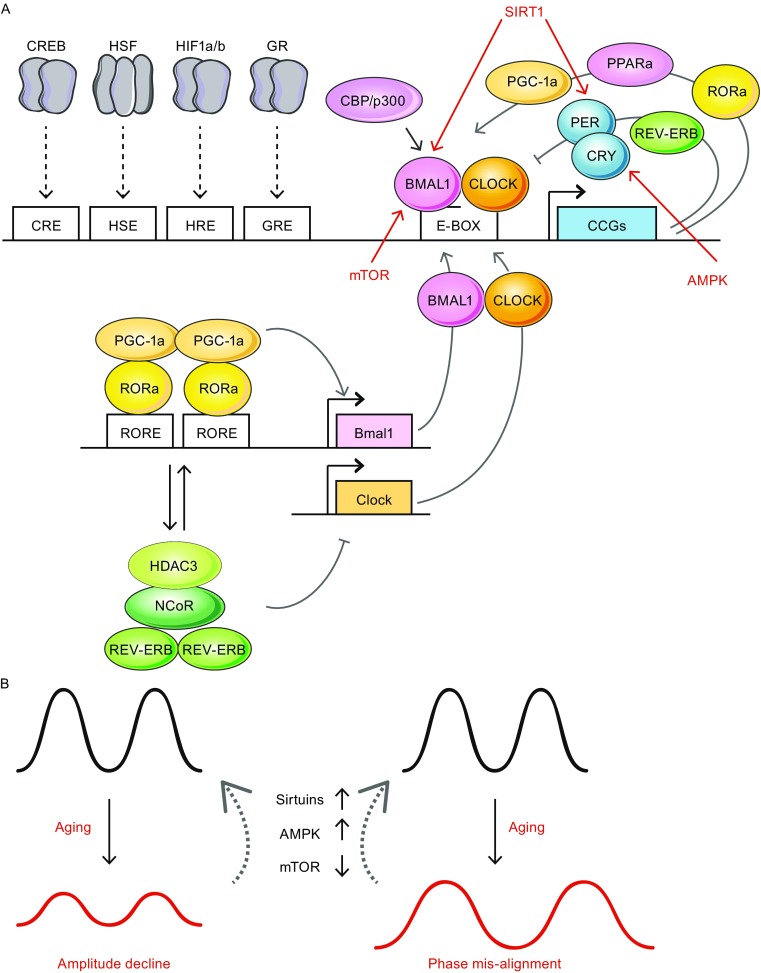
Molecular oscillators in circadian control. (A) Transcription factor complex CLOCK:BMAL1 binds to E-box containing motifs, allows the transcriptional activation of clock-controlled genes (CCGs) such as Pers, Crys, Ror and Rev-Erb. The activation is facilitated by recruiting coactivators such as CBP/p300. CCG transcriptions are as well regulated by transcription factors relaying the external cues. Examples include cAMP responsive element binding protein (CREB), heat shock factor 1 (HSF1), hypoxia-inducible factor 1α (HIF1α) and glucocorticoid receptor (GR) that bind to their respective regulatory elements (Bollinger and Schibler, 2014; Wu et al., 2016). Two interconnected feedback loops involved in the circadian transcriptional regulation. In the primary feedback loop, PER and CRY assemble into repressor complexes next attenuate the activity of CLOCK:BMAL1. In the second feedback loop, ROR (also a CCG protein) can complex with coactivator PGC-1α and bind to RORE element for Bmal1 (and likely Clock) activation(s). REV-ERB works as a repressor in Bmal1 transcription by concentration-dependent competition at the same RORE sequence. The repression involved the recruitment of NCoR/HDAC3 corepressor complexes. (B) Energy sensors such as Sirtuins, AMPK and mTOR participate in circadian modulations via post-translational modification of circadian components, as depicted in (A). Interventions target the pathways are of potential to treat age-associated circadian amplitude decline and phase mis-alignment
