Figure 1.
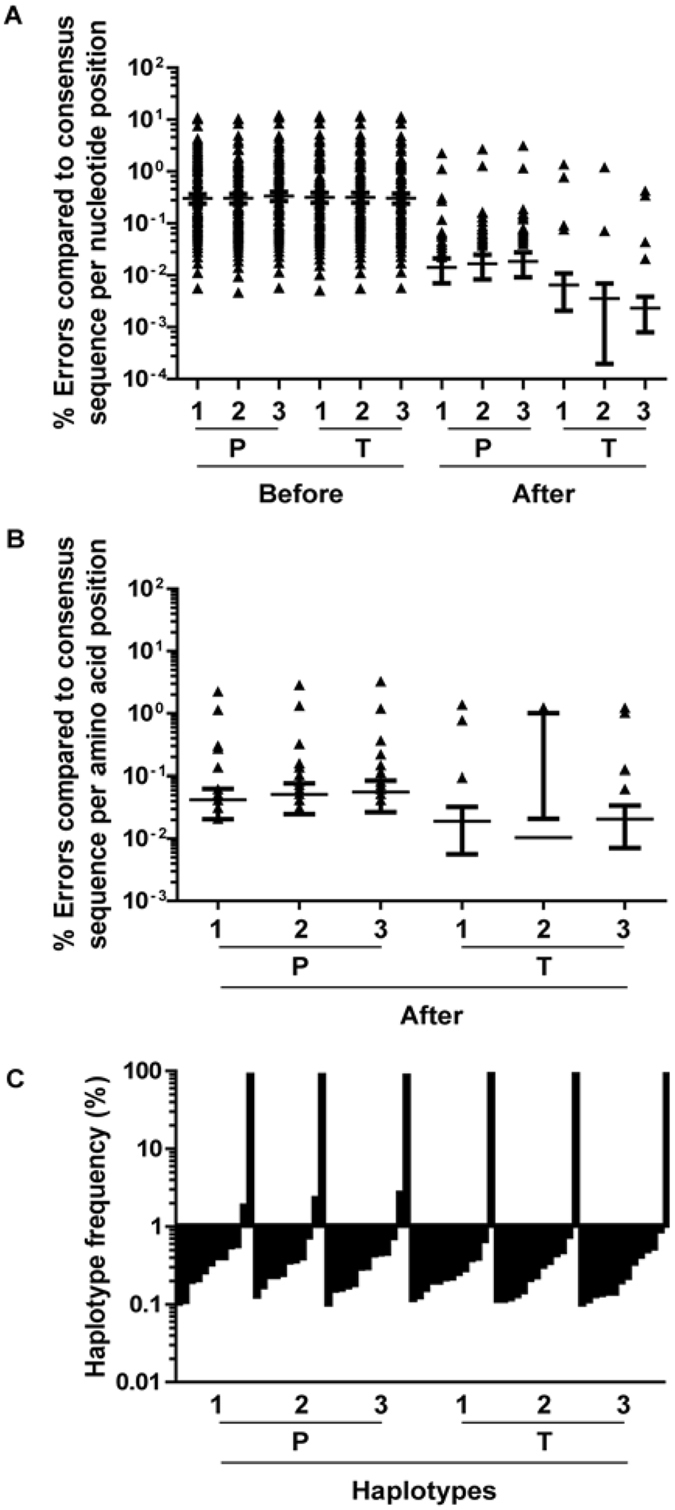
Validation of the next-generation sequencing assay. (A) A position-specific error rate per nucleotide was determined before or after read-cleaning algorithms were applied for the control experiments with plasmid (P) or RNA transcript (T) in 3 independent next-generation sequencing experiments (1–3) each. Mean ± sem are shown. (B) For confirmation, a position-specific error rate per amino acid position was determined after read-cleaning algorithms were applied for the control experiments with plasmid (P) or RNA transcript (T) in 3 independent next-generation sequencing experiments (1–3) each. Mean ± sem are shown. (C) Haplotypes and their frequencies in the dataset after read-cleaning were determined for the control experiments with plasmid (P) or RNA transcript (T) in 3 independent next-generation sequencing experiments (1–3) and plotted against each other with a cut-off value of 0.1%.
