Figure 1.
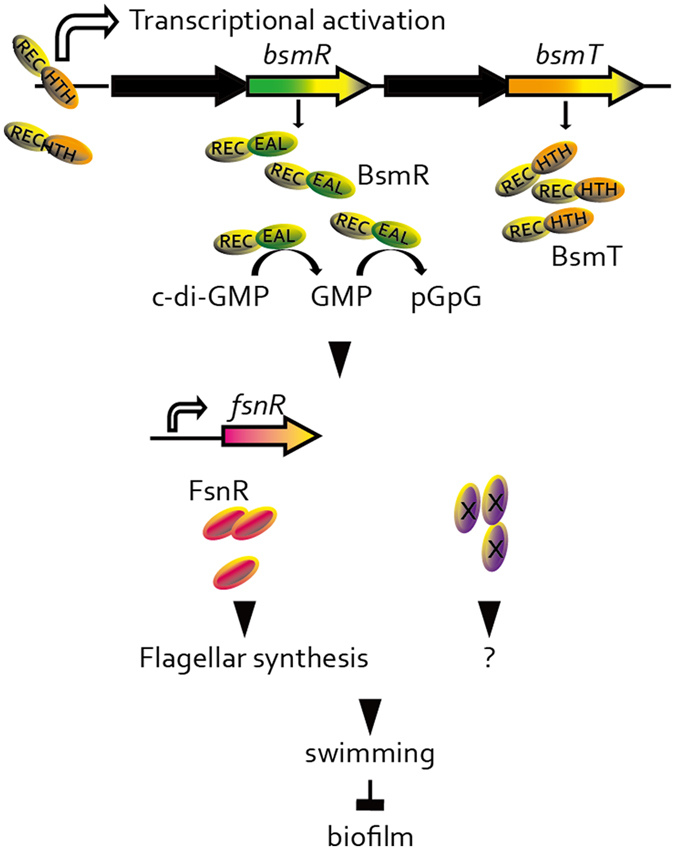
Working model. BsmR degrades c-di-GMP to decrease its cellular level, which activates the expression of FsnR, a transcriptional regulator controlling flagellar synthesis, to promote bacterial swimming motility and diminish the biofilm formation. Although as a response regulator, the PDE activity and the regulatory role of BsmR on bacterial swimming motility and biofilm formation do not depend on the phosphorylation level of BsmR but on its expression level, which is controlled by BsmT, another response regulator encoded by the bsmR operon. BsmT, working as a transcriptional regulator, directly binds to the promoter region of the bsmR operon and positively regulates the expression of all four genes in the bsmR operon, including bsmR. REC: conserved receiver domain; HTH: helix-turn-helix DNA binding domain; EAL: EAL domain; hallow arrows: transcription activation; ×: unknown regulatory elements;?: unknown regulatory role of ×.
