Fig. 3.
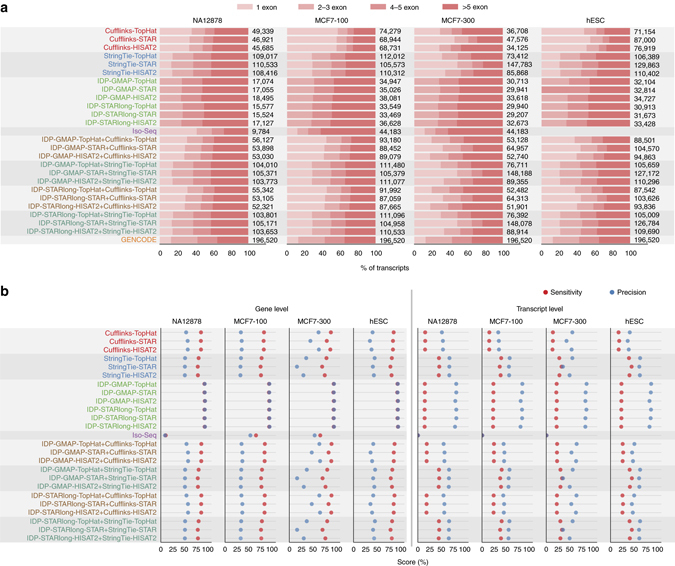
Performance of different transcriptome reconstruction schemes. a Distribution of number of exons per transcripts for different transcriptome reconstruction algorithms. Labels reflect the assembler, the long-read aligner (for IDP), and the short-read aligner used, respectively, with “-” separation. b Sensitivity and precision of different transcriptome reconstruction approaches at gene and transcript levels. The GENCODE reference transcriptome annotation is used as the truth set. The evaluations on a more recent update of MCF7 sample using the Iso-Seq pipeline resulted in a similar performance with only slight improvement. The union approaches that combined predictions from short reads and long reads (shown with a “+” in the label) slightly improved the performance of short-read isoform prediction schemes
