Fig. 3.
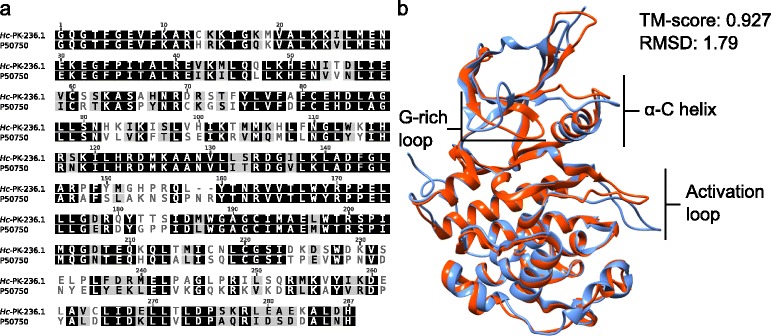
Multiple-sequence alignment showing levels of similarity in the kinase catalytic domains (Pfam identifier: PF00069) of CDK9 homologs between Haemonchus contortus (Hc-PK-236.1) and human (UniProt accession no.: P50750). a The pairwise sequence alignment was constructed using the program MUSCLE [39]. b Three-dimensional model of CDK9 homolog of H. contortus (Hc-PK-236.1; orange) superimposed on to the crystal structure of human CDK9 (protein data bank (PDB) identifier: 4or5A; blue). The TM-score and root-mean-square deviation (RMSD) values indicate a high-confidence prediction. Conformational differences predicted in G-rich loop, activation loop and α-C helix between the two structures are indicated. The three-dimensional structure for Hc-PK-236.1 was predicted using the program I-TASSER [40] using default parameters
