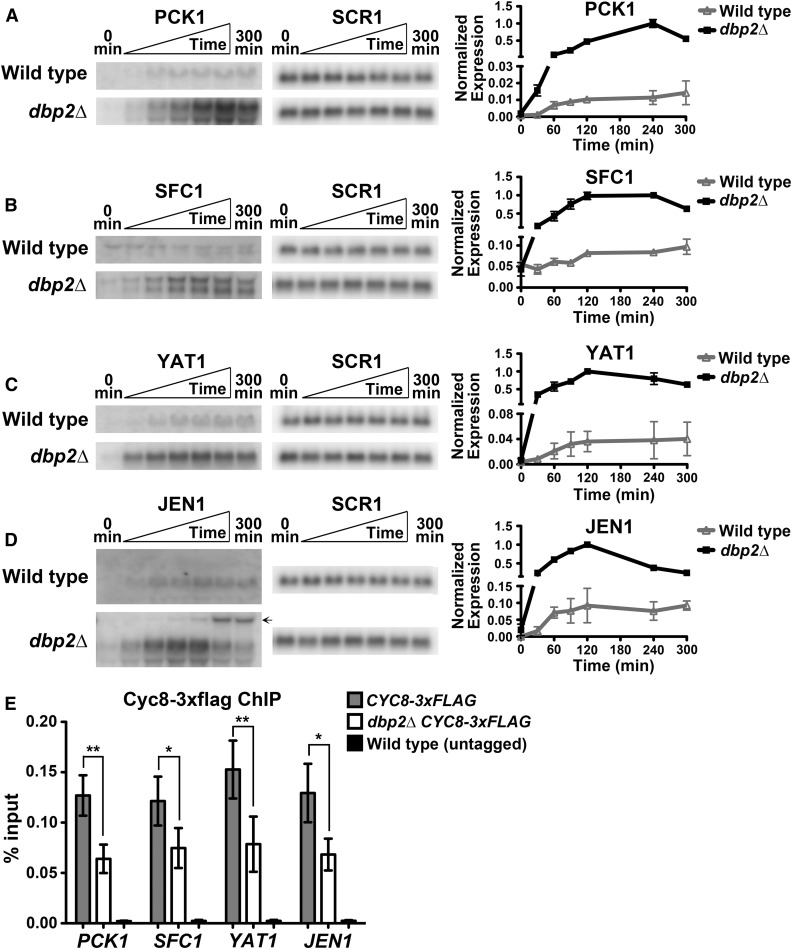
Figure 5.
Loss of DBP2 results in reduced Cyc8 binding and rapid induction of genes involved in utilizing nonfermentable carbon source. (A–D) Transcriptional induction assays of PCK1 (A), SFC1 (B), YAT1 (C), and JEN1 (D) were conducted by shifting wild-type and dbp2∆ cells from SC+2% glucose to SC+1% potassium acetate media. RNA was collected from cells prior to (0 min) and at 30, 60, 90, 120, 240, and 300 min following nutritional shift. Transcripts were detected by Northern blotting using 32P-labeled, dsDNA probes. SCR1 serves as an internal control. Resulting transcript profiles from two biological replicates (error bars indicate ± SD) were normalized to SCR1 and plotted over time (right). The normalized expression of the time points with maximum transcript levels in each induction profile was set to one. The transcript, presumably XUT1417, is labelled ←. (E) ChIP of Cyc8-3xFLAG in wild-type and dbp2∆ cells. Wild-type and dbp2∆ strains harboring 3xFLAG-tagged CYC8 at the endogenous locus were cultivated in YP+2% glucose. ChIP was conducted using an anti-FLAG antibody. A wild-type strain with untagged CYC8 was used as negative control. Results were represented as percentage of immunoprecipitated DNA over input and shown as mean ± SD of three independent biological replicates. *P < 0.05, **P < 0.01.