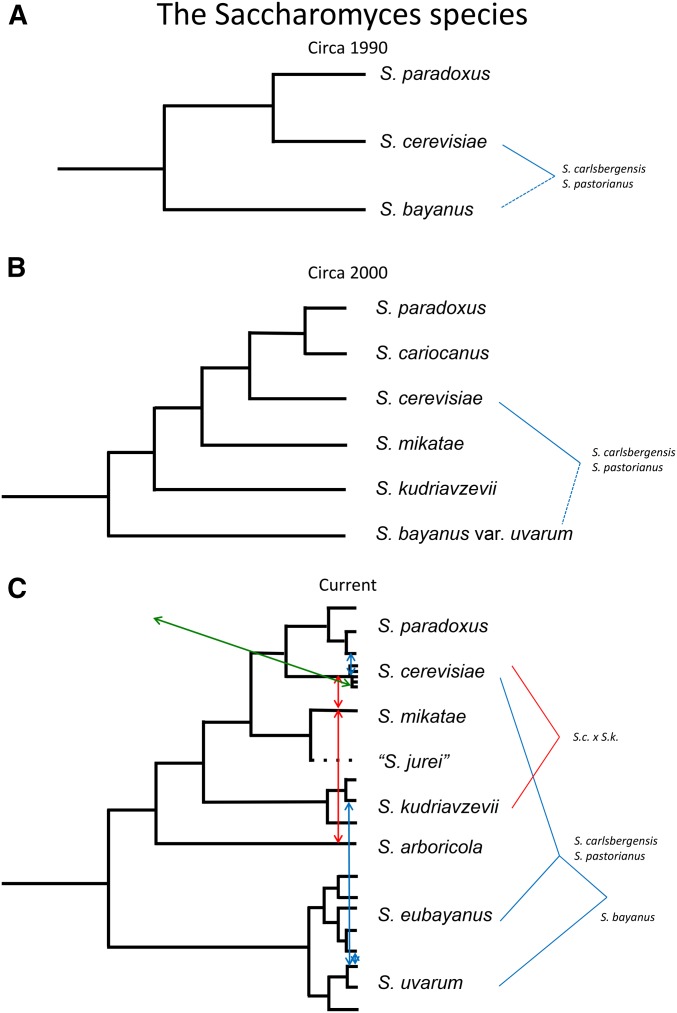
Figure 1.
Evolution of the phylogeny of the Saccharomyces (formerly S. sensu stricto) group since the first genome sequence. (A) By the mid-1980s to mid-1990s, the use of DNA–DNA reassociation (Vaughan Martini and Martini 1987; Vaughan Martini 1989) and the biological species definition (Naumov 1987) led to the consolidation of the Saccharomyces yeasts into three species and one hybrid used in lager fermentation. This hybrid was between S. cerevisiae and something close to S. bayanus, but not S. bayanus itself (Casey and Pedersen 1988; Hansen and Kielland-Brandt 1994; Hansen et al. 1994). (B) By the late 1990s, the use of the biological species definition, along with electrophoretic karyotyping and presence/absence of specific repeated sequences, on isolates in various culture collections resulted in the discovery of three new species, S. cariocanus, S. mikatae, and S. kudriavzevii, and the refinement of S. bayanus var. uvarum as a species while S. bayanus itself appears to be a hybrid. (C) In recent years, whole genome sequencing along with genetic analysis has resulted in the current view of the group. One species (S. cariocanus) has disappeared based on phylogeny (see text) and three others have been discovered (S. arboricola; S. eubayanus, the other parent in the lager hybrids; and S. jurei). There are many examples of HGT, red arrows, as well as introgressions, blue arrows (see text). Perhaps the most interesting is the HGT of genes that provide useful traits in wine fermentation, green arrow, which distinguishes the wine yeast from wild European yeasts (see text and Fig. 2).