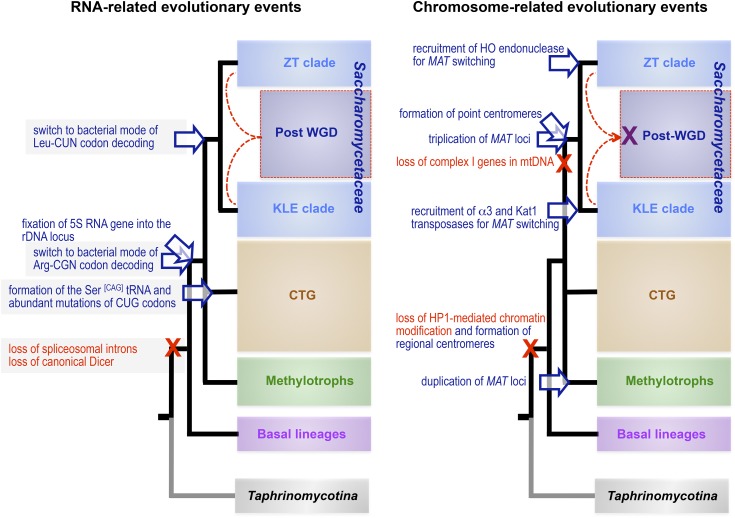
Figure 9.
Overview of Saccharomycotina genome evolution. Evolutionary innovations (blue arrows) or losses (red crosses) attributed from parsimony to major separations between subgroups of Saccharomycotina (secondary losses of preceding innovations, not shown for simplicity). Left: RNA-related events. Note that, in some lineages, noncanonical enzymes replaced the ancestral Dicer and that the phylogenetic distribution of Dicer and Argonaute proteins in Saccharomycotina is discontinuous (see text). Right: chromosome-related events. Note that the recruitment of α3 and Kat1 transposases concerns the Kluyveromyces, not the entire KLE clade, and that the loss of H3K9me chromatin modification occurred after the separation of L. starkeyi (data not shown) from other basal lineages. Purple cross: massive post-WGD gene loss. Note that the loss of complex-I genes in mtDNA is not a chromosomal event, but its occurrence is indicated because of its association with the duplication of nuclear genes encoding alternative dehydrogenases (see text).