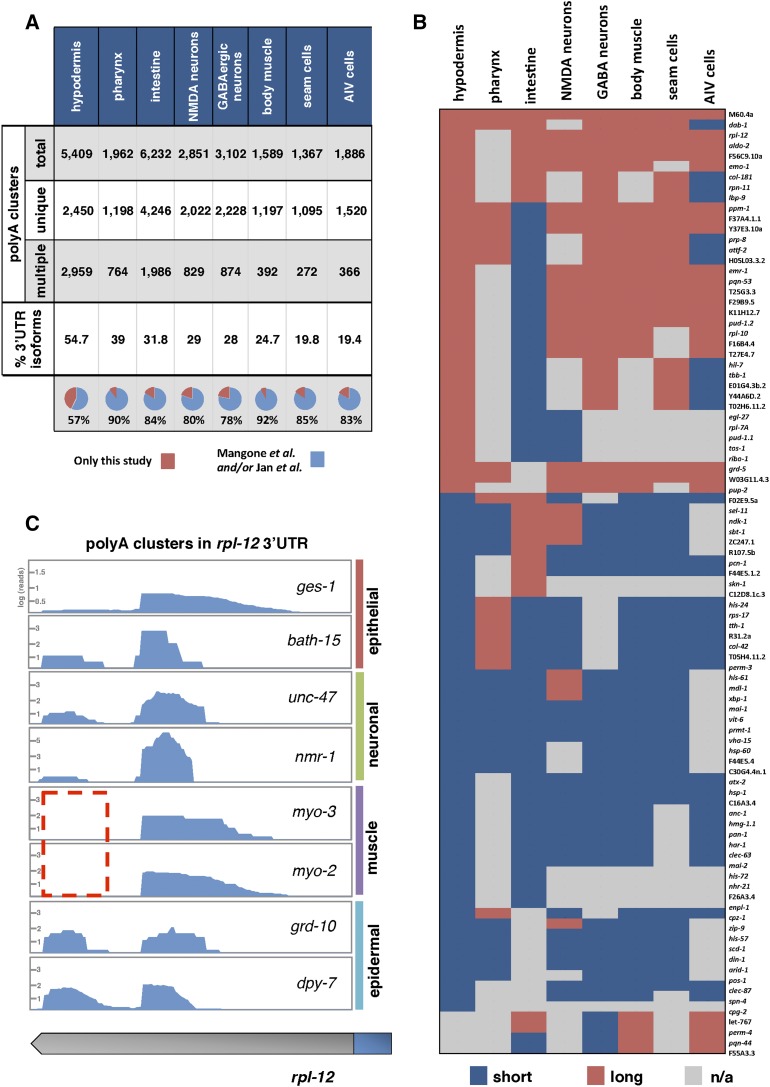
Figure 3.
PolyA Cluster mapping. (A) We have bioinformatically assembled ∼25,000 high-quality polyA clusters distributed in eight somatic C. elegans tissues from our RNA-seq data. These clusters map the 3′ends of protein coding genes. Genes in these tissues use APA at a ∼31% rate; >83% of these mapped polyA clusters map 3′UTR isoforms previously detected by Mangone et al. (2010) and Jan et al. (2011) (B) Tissue-specific 3′UTR isoform preferences in 91 protein coding genes detected with only two 3′UTR isoforms in this study. Most of the genes in this analysis use tissue-specific APA. Instances where a 3′UTR was not mapped in a tissue are indicated in gray (n/a). (C) Example of polyA clusters prepared for the gene rpl-12. The distal 3′UTR isoform is absent in both muscle tissues (dotted red box).