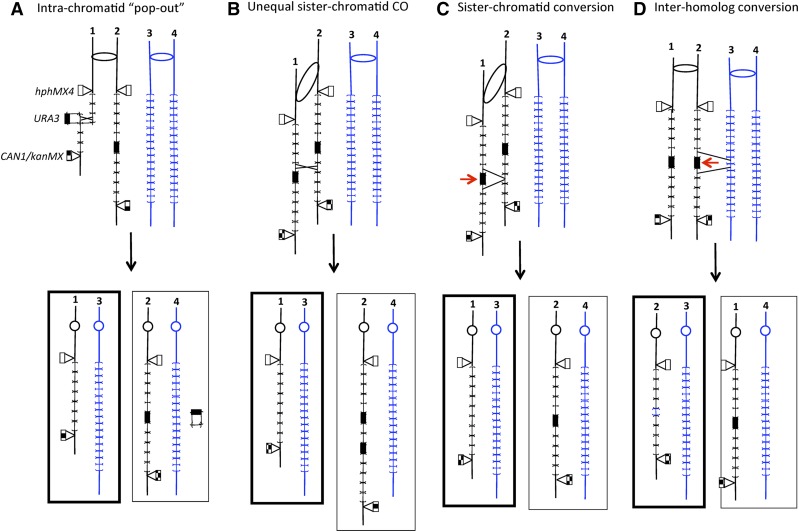
Figure 2.
Mitotic recombination events leading to loss of URA3 marker and retention of the flanking hphMX4 and CAN1/kanMX4 markers. The chromosomes are shown following DNA replication, and the two homologs are shown in different colors; in this depiction, one homolog has 2-kb CUP1 repeats and the other 1.2-kb repeats. CUP1 repeats are indicated by brackets. The chromosomes of 5-FOAR daughter cells are outlined by thick lines. (A) Intrachromatid “pop-out” recombination. A crossover occurs within a chromatid, producing a shorter CUP1 array and a plasmid with the URA3 gene and three CUP1 repeats. The cell with the chromosomes outlined with thick lines would be 5-FOAR. Since each CUP1 repeat has an ARS element, the URA3-containing plasmid would be capable of autonomous replication. It is shown segregated into the daughter cell that also contains an integrated URA3 gene. (B) Unequal sister chromatid crossover. As a consequence of this event, the 5-FOAR daughter cell would contain a shorter CUP1 array, and the Ura+ daughter cell would contain a longer array with two URA3 insertions. (C) Intersister chromatid gene conversion. A DSB (shown as a red arrow) occurs near the URA3 insertion in one chromatid, and is repaired using the sister chromatid as a template. The net result of this event would be a loss of URA3 and one or more CUP1 repeats in one daughter cell with no alteration in the second daughter. (D) Interhomolog gene conversion. As in (C), this event is initiated with a DSB near or within the URA3 insertion. The repair template, however, is a chromatid of the other homolog instead of the sister chromatid. Associated with the loss of URA3 and the loss of some of the 2.0-kb CUP1 repeats, one would expect insertion of one or more 1.2-kb CUP1 repeats derived from the other homolog. 5-FOA, 5-fluoroorotic acid; CO, crossover; DSB, double-stranded DNA breaks; R, resistant; Ura+, uracil+.