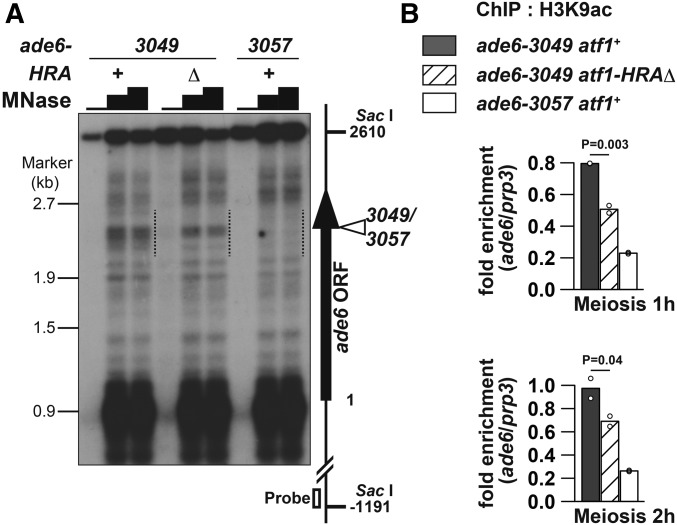
Figure 5.
Chromatin structure around ade6-3049 is more open than around ade6-3057, and HRA is involved in the chromatin regulation. (A) MNase sensitivity of chromatin around ade6-3049/3057. Indicated cells of the pat1-114 background were induced to enter meiosis by temperature-shift method, and harvested 3 hr after the induction. MNase-digested DNA was prepared by MNase (0, 20, or 30 units) treatment of chromatin fraction and subsequent purification. DNA was further cut with Sac I, fractionated on 1.2% agaraose gel, and analyzed by Southern blotting using the probe indicated by the open box. The vertical arrow and the open arrowhead show the ade6 ORF and the 3049/3057 site, respectively. The numbers on the left and the right side of the panel indicate the positions of λ/EcoT14 I fragments used for electrophoresis marker and the positions from the first A of the ade6 ORF, respectively. The dotted lines indicate a region in which MNase-sensitive sites are clustered around ade6-3049. Presented is an example from two independent experiments, whose results were similar to each other. (B) HRA deletion reduced acetylation level of H3K9 around ade6-3049. Indicated cells of the pat1-114 background were induced to enter meiosis by temperature-shift method, and harvested 1 or 2 hr after the induction. ChIP using anti-H3K9ac antibody was performed as described previously (Yamada et al. 2013). DNA isolated from immunoprecipitates and whole-cell extracts was analyzed by real-time qPCR, where fragments corresponding to ade6-3049/3057 and the prp3+ promoter were amplified. Relative enrichment at ade6-3049/3057 over prp3 is shown. Bar graphs are created based on mean values of two independent experiments (shown by ○).