Abstract
Oxidative damage contributes to human diseases of aging including diabetes, cancer, and cardiovascular disorders. Reactive oxygen species resulting from xenobiotic and endogenous metabolites are sensed by a poorly understood process, triggering a cascade of regulatory factors and leading to the activation of the transcription factor Nrf2 (Nuclear factor-erythroid-related factor 2, SKN-1 in Caenorhabditis elegans). Nrf2/SKN-1 activation promotes the induction of the phase II detoxification system that serves to limit oxidative stress. We have extended a previous C. elegans genetic approach to explore the mechanisms by which a phase II enzyme is induced by endogenous and exogenous oxidants. The xrep (xenobiotics response pathway) mutants were isolated as defective in their ability to properly regulate the induction of a glutathione S-transferase (GST) reporter. The xrep-1 gene was previously identified as wdr-23, which encodes a C. elegans homolog of the mammalian β-propeller repeat-containing protein WDR-23. Here, we identify and confirm the mutations in xrep-2, xrep-3, and xrep-4. The xrep-2 gene is alh-6, an ortholog of a human gene mutated in familial hyperprolinemia. The xrep-3 mutation is a gain-of-function allele of skn-1. The xrep-4 gene is F46F11.6, which encodes a F-box-containing protein. We demonstrate that xrep-4 alters the stability of WDR-23 (xrep-1), a key regulator of SKN-1 (xrep-3). Epistatic relationships among the xrep mutants and their interacting partners allow us to propose an ordered genetic pathway by which endogenous and exogenous stressors induce the phase II detoxification response.
Keywords: C. elegans, stress response, detoxification, XREP
OXIDATIVE stress is widely recognized to be a major contributor to the pathophysiology of numerous diseases including diabetes, cancer, and cardiovascular and neurodegenerative disorders. The downstream defense mechanisms providing protection against reactive oxygen species, a major source of acute oxidative stress, are mediated by a highly-conserved set of phase II detoxification enzymes including the glucuronosyltransferases and the glutathione S-transferases (GSTs) (Jakoby and Ziegler 1990). These enzymes act in combination to metabolize almost any hydrophobic compound that contains nucleophilic or electrophilic groups. Toxic compounds generated by normal metabolism or because of phase I detoxification of xenobiotics are primarily acted upon by the glutathione transferases, facilitating their removal. Thus, cellular detoxification mechanisms must sense oxidative or xenobiotic insults resulting from a wide range of endogenous and exogenous stimuli, which activate a battery of cellular response genes with broad specificity and high capacity.
Caenorhabditis elegans, like mammals, exhibits evolutionarily conserved mechanisms for dealing with cellular stress, including the MAPK kinase cascades, insulin signaling, and nuclear factor-erythroid-related factor (Nrf)/SKN-1 pathways regulating genes that encode GSTs (Carroll et al. 1997; Pal et al. 1997; Rupert et al. 1998; Kahn et al. 2008; Hasegawa and Miwa 2010; Sykiotis and Bohmann 2010; Li et al. 2011; Paek et al. 2012; Glover-Cutter et al. 2013; Pang et al. 2014; Blackwell et al. 2015). C. elegans provides an unbiased genetic means of identifying important regulatory components in these signaling and transcriptional pathways. One common assay is the activation of a gst-4 reporter gene, used previously to identify genes involved in the response to acrylamide (Hasegawa et al. 2008), cadmium (Roh et al. 2009), and other sources of oxidative stress (Hasegawa et al. 2007, 2010; Hasegawa and Miwa 2010; J. Wang et al. 2010; Jones et al. 2013; Leung et al. 2013; Crook-McMahon et al. 2014).
In a forward genetic screen for acrylamide-responsive genes (Hasegawa and Miwa 2010), a gst-4::gfp reporter was used to identify a collection of xenobiotics response pathway (xrep) mutants. Of the 24 mutants identified in this screen, four complementation groups were reported (xrep-1, -2, -3, and -4). The xrep-1 gene was identified as wdr-23, the nematode homolog of the mammalian β-propeller repeat-containing protein WDR-23 (Hasegawa and Miwa 2010). Prior evidence indicated that gst-4 expression was regulated in part by SKN-1 (Hasegawa et al. 2008). In mammalian systems, the β-propeller repeat protein Keap1 interacts with Nrf2, the ortholog of SKN-1, to govern oxidative stress response genes (Itoh et al. 1999; Kobayashi et al. 2004; Osburn and Kensler 2008; Nguyen et al. 2009). A functional equivalence was proposed for WDR-23 and SKN-1 in the regulation of acrylamide-responsive genes in C. elegans (Choe et al. 2009; Przybysz et al. 2009; Hasegawa and Miwa 2010); the molecular identities of the remaining xrep mutations remained to be determined.
In this report, we have employed whole-genome sequencing (WGS) with Hawaiian SNP mapping (Doitsidou et al. 2010), candidate gene sequencing, RNAi phenocopy, transgenic assays, and mutant rescue to identify xrep-2, -3, and -4. The xrep genes alh-6 (xrep-2), skn-1 (xrep-3), and the F-box protein-encoding gene F46F11.6 (xrep-4), in conjunction with the previously identified wdr-23 (xrep-1), form a coherent genetic signaling pathway based on epistasis analysis. These results provide a framework for understanding the organismal response to endogenous and exogenous oxidative stress, and support the increasingly widespread use of C. elegans as a model for toxicology and high-throughput drug screening (Hasegawa et al. 2004, 2007; Leung et al. 2013; Rangaraju et al. 2015).
Materials and Methods
Strains and cultures
Standard C. elegans culture conditions were used (Brenner 1974). The following strains were used in this study: N2 (Bristol), CB4856 (wild-type, Hawaiian), MJCU017 (unc-119(ed3) III, kIs17[gst-4::gfp, pDPMM#016B] X) referred to throughout as gst-4::gfp, MJCU047 (unc-119(ed3) III, kIs41[gst-30::gfp, pDPMM#016B] X) referred to throughout as gst-30::gfp, MJCU085 (unc-119(ed3) III, kIs84[xrep-1(+)::gfp, pDP#MM016B]) referred to throughout as wdr-23::gfp, MJCU1007 wdr-23(k1007); gst-4::gfp, MJCU1018 alh-6(k1018); gst-4::gfp, MJCU1022 alh-6(k1022); gst-30::gfp, MJCU1023 skn-1(k1023); gst-4::gfp, and MJCU1024 xrep-4(k1024); gst-4::gfp. Isolation of the xrep mutants was previously described (Hasegawa and Miwa 2010). Acrylamide exposure used NGM plates containing 200 mg/liter of acrylamide.
Mutation identification
The xrep-2 and xrep-4 mutations were identified by WGS (Table 1). Mutation intervals were determined by the one-step SNP mapping method (Doitsidou et al. 2010) via crosses to Hawaiian strain CB4856 (Hodgkin and Doniach 1997). Libraries from each strain were constructed using either NEBNext DNA or Ultra DNA library prep kits for Illumina (Cat. Nos. E6040 or E7370, respectively, New England Biolabs, Beverly, MA). Single-end 50 bp sequencing was performed on a HiSequation 2500 instrument (Illumina, San Diego, CA), yielding a minimum of 20-fold genome coverage for each library. Variants were identified using a pipeline of BBMap for alignment (Bushnell 2015), FreeBayes for variant calling (Garrison and Marth 2012), ANNOVAR for gene annotation (K. Wang et al. 2010), BEDTools for Hawaiian SNP annotation (Quinlan and Hall 2010), and R for Hawaiian SNP frequency plots (R Core Team 2016). Candidate mutations were defined as nonparental, homozygous, and nonsynonymous variants within the map interval (Table S1). The gain-of-function skn-1(k1023) allele, previously identified as xrep-3(k1023), was determined by Sanger sequencing of the skn-1 exons amplified from the strain MJCU1023.
Table 1. Strains for whole-genome sequencing.
| Strain | Description | Mutationa | Gene and substitutionb |
|---|---|---|---|
| MJCU017 | gst-4::gfp parental strain | N/A | N/A |
| K1017 | xrep-2; gst-4::gfp | ChrII: 1,306,476, C > T | F56D12.1, Gly534Asp |
| K1018 x CB4856 | xrep-2; gst-4::gfp Haw cross | ChrII: 1,306,476, C > T | F56D12.1, Gly534Asp |
| K1019 | xrep-2; gst-4::gfp | ChrII: 1,306,476, C > T | F56D12.1, Gly534Asp |
| K1020 | xrep-2; gst-4::gfp | ChrII: 1,306,476, C > T | F56D12.1, Gly534Asp |
| K1021 | xrep-2; gst-4::gfp | ChrII: 1,306,476, C > T | F56D12.1, Gly534Asp |
| K1022 | xrep-2; gst-30::gfp | ChrII: 1,306,500, G > A | F56D12.1, Ser526Phe |
| K1024 | xrep-4; gst-4::gfp | ChrI: 5,617,911, C > T | F46F11.6, Arg92Opal |
N/A, not applicable; Chr, chromosome.
Mutation position based on reference genome version WS250 (www.wormbase.org).
Amino acid position based on isoforms F56D12.1a and F46F11.6a, respectively.
RNAi constructs and procedures
The xrep-2 mutation was confirmed as alh-6 via RNAi phenocopy by injecting alh-6 dsRNA into the gst-4::gfp translational fusion reporter strain. To identify the xrep-4 mutation, 12 of 26 genes (unc-89, cec-10, F27C1.3, F46F11.6, tyr-4, C48E7.6, hrpk-1, pcbd-1, dcp-66, apb-3, mys-4, and B0511.12) in the Hawaiian SNP mapping interval by WGS were tested individually by injecting dsRNA into the MJCU1018 (alh-6 mutant) strain. To test SKN-1 dependence of GST activation, a skn-1-specific RNAi construct was used that does not include any conserved nucleotide sequence with the related gene, sknr-1. The skr-1/2 RNAi construct used the skr-1 gene as a template for cDNA amplification, which is ∼83% identical to skr-2 at the nucleotide level. All RNAi clones were generated by amplifying target sequences using a wild-type cDNA preparation. Gel-purified amplicons were inserted into the L4440 plasmid that was used to synthesize dsRNA. Most of the RNAi experiments were performed by injection of dsRNA into the gonads of adult animals using standard techniques and assaying the progeny. In some cases, RNAi knockdown of gene function was achieved by feeding RNAi (Ahringer 2006) starting with L1-stage animals. Primers used for all RNAi constructs are shown in Supplemental Material, Table S2.
Mutant rescue
All injections to generate transgenic strains included the dominant rol-6(su1006) plasmid (pRF4, 100 ng/μl) as a visible marker (Mello et al. 1991). To rescue mutants, genomic regions encompassing either wild-type alh-6 or xrep-4 were amplified by PCR with alh-6pF1 and alh-6R(3′UTR) or F46F11.6pF and F46F11.6R(3′UTR) primer sets, respectively. Purified PCR fragments (1 ng/μl) were injected into corresponding alh-6(k1018) or xrep-4(k1024) mutant strains. Plasmids of the genomic fragments used for rescue were constructed with a C-terminal mCherry tag so that the rescuing protein products could be visualized. Amplified genomic DNA fragments were cloned into pCR2.1-TOPO by using the TOPO cloning kit (Invitrogen, Carlsbad, CA, cat#K4500-01). An mCherry::unc-54 3′-UTR fragment from pKM1271 was inserted into the 3′-end of the alh-6 or xrep-4 gene construct via standard cloning methods. The appropriate mCherry-tagged rescue construct (10 or 40 ng/μl) was injected into either alh-6(k1018) or xrep-4(k1024) mutant strain and transgenic progeny were scored for their gst-4::gfp expression phenotypes.
For xrep-4 mutants, rescue following tissue-restricted expression of the wild-type xrep-4 genomic region was tested with promoters driving expression in the intestine [pho-1 promoter (pKM1272)] or muscle [myo-3 promoter (pKM1273)]. The tissue-specific constructs (50 ng/μl) were injected into the gst-4::gfp strain MJCU017. Detailed primer information is provided in Table S2.
Imaging and processing
Animals were mounted either on agarose pads or anesthetic buffer solution (100 μM levamisole in PBS) and imaged using a Nikon CFI60 microscope system (Nikon, Garden City, NY) fitted with a Retiga 2000R digital camera (Qimaging) and captured with iVision imaging software (BioVision Technologies). Image data were processed using Adobe Photoshop CC software.
Western analysis
Synchronized L1 animals were prepared from wdr-23::gfp and wdr-23::gfp; xrep-4(k1024) populations. L1 worms were grown on NGM plates for ∼24 hr at room temperature after recovering from starved conditions used to synchronize the L1 population. Animals were transferred to fresh plates with or without acrylamide. Animals were incubated for another ∼24 hr at room temperature before total protein was extracted at the late L3 and early L4 stages using a Mini-Beadbeater-16 (Biospec Products). Protein concentrations were measured by 280 nm absorbance (Nanodrop 2000c, Thermo Scientific) and similar amounts of protein loaded for gel electrophoresis and western blotting. The anti-GFP antibody (A-11120, Thermo Scientific) was used to detect WDR-23::GFP and the anti-α-tubulin antibody (DM1A, Sigma [Sigma Chemical], St. Louis, MO) was used to detect α-tubulin as the internal loading and transfer control. The anti-mouse peroxidase (Cat# 715-035-150, Jackson ImmunoResearch) was used as a secondary antibody and the SuperSignal West Dura Extended Duration Substrate (Cat# 34075, Thermo Scientific) was used for detection of the signal. Signals were captured using the digital gel imaging system “Fluorchem E” (ProteinSimple) and the results averaged over at least three biological replicates for each strain and growth condition.
Data availability
Strains and genomic sequences are available upon request. All oligonucleotides used for cloning are listed in Table S2.
Results
XREP-2 is ALH-6, linking aberrant proline catabolism to the constitutive stress response
The initial genetic mutant screen recovered six xrep-2 strains that exhibit constitutive adult expression of gst-4::gfp (five strains) or gst-30::gfp (one strain) reporter genes in the absence of acrylamide exposure (Hasegawa and Miwa 2010); gst-4::gfp expression was most evident in bodywall muscle (BWM) whereas gst-30::gfp was strongest in the posterior pharyngeal bulb. We utilized WGS with the one-step Hawaiian SNP mapping method of one representative xrep-2 strain (MJCU1018) to delimit the mapping interval to chromosome II, between 0 and 2 Mb (Doitsidou et al. 2010). We also performed WGS and variant calling for the remaining xrep-2 strains and the unmutagenized parental strain that contained the gst-4::gfp translational reporter. Comparisons of de novo (nonparental) variants revealed that the five strains derived from the gst-4::gfp-containing parent were genetically identical and, likely, represented a single clonal line rather than independently generated alleles; a single independent mutant, xrep-2(k1022), was isolated in the gst-30::gfp reporter background. By comparing xrep-2 candidate mutations from the gst-4::gfp and gst-30::gfp-bearing strains we identified a single gene, alh-6, that had distinct mutations in the two strains.
The alh-6 gene encodes an aldehyde dehydrogenase most similar to the human aldehyde dehydrogenase 4 family (ALDH4A1). These are highly conserved, NAD-dependent enzymes found in the mitochondrial matrix that catalyze a step in the proline degradation pathway; loss of ALDH4A1 activity in humans results in recessive type II hyperprolinemia disorder (Flynn et al. 1989). ALDH4A1 converts δ-1-pyrroline-5-carboxylate (P5C) to glutamate and its loss leads to the accumulation of P5C, which is toxic to cells and tissues (Mitsubuchi et al. 2008). Interestingly, a previous genetic screen in C. elegans has also linked alh-6 proline catabolism to lipid oxidation and the phase II detoxification response mediated by SKN-1 (Pang and Curran 2014; Pang et al. 2014).
The mutation we identified in the xrep-2(k1018) strain (and clonal isolates) was in the last exon of the alh-6a isoform, resulting in a Gly534Asp amino acid substitution (Figure 1A). The mutation in the xrep-2(k1022) strain was similarly located in the last exon of alh-6, resulting in a Ser526Phe substitution (Figure 1A). Both mutations alter the evolutionarily conserved C-terminal domain of ALH-6a. Structures of the highly homologous mouse and human ALDH4A1 proteins localized the binding sites for NAD and glutamate to this domain (Srivastava et al. 2012). Mutations that we (Figure 1A, in blue) and others (in black) have identified in alh-6a are adjacent to the active site residues that contact glutamate substrate (in red) in mouse ALDH4A1; this active site is also near the conserved NAD-binding site. These findings demonstrate that many of the mutations identified to date in ALH-6a map to the region corresponding to the conserved NAD/glutamate-interaction domain.
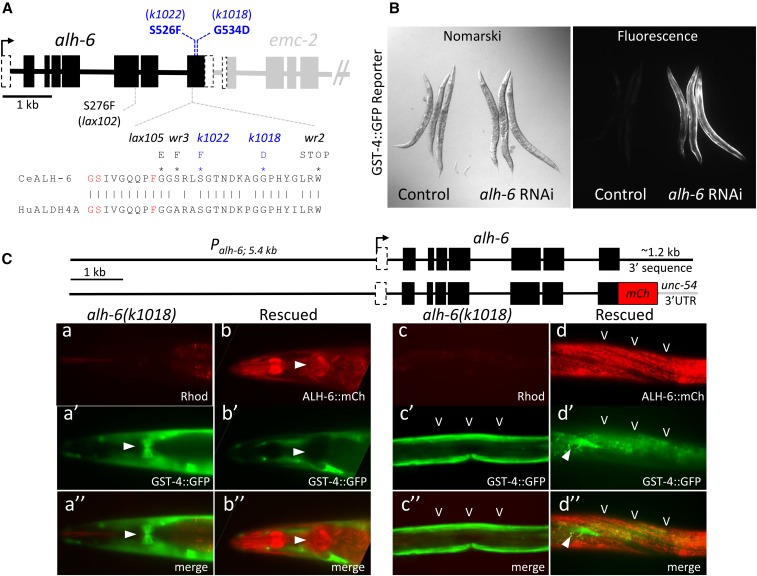
Figure 1.
Identification of xrep-2 as an allele of alh-6. (A) The alh-6 gene structure and mutations. The gene structure of alh-6 is diagramed (black) along with part of its neighboring gene in the operon, emc-2 (gray). The positions of alleles identified in this study (k1018 and k1022) are shown in blue relative to several previously identified mutations [Schlipalius et al. (2012) and Pang and Curran (2014)]. Many cluster in the last exon, which encodes an evolutionarily conserved interface between the inferred substrate and NAD+-binding pockets of ALH-6 based on sequence homology to mammalian ALDH4A1 (Srivastava et al. 2012). A segment of the protein sequence from this region is shown, with active site residues in red and mutant substitutions (black and blue) as indicated. (B) Phenocopy of the xrep-2 mutation by alh-6 RNAi (RNA interference). Control wild-type adult animals harboring the gst-4::gfp translational fusion reporter gene are shown next to the same strain after alh-6 RNAi. Knockdown of alh-6 activity results in strong upregulation of gst-4::gfp expression in bodywall muscles (BWMs). (C) alh-6 mutant rescue. Genomic wild-type and mCherry (mCh)-tagged alh-6-rescuing constructs are diagramed at the top; each was introduced separately into alh-6(k1018) mutants harboring the gst-4::gfp reporter gene and stable extrachromosomal strains were established. The left panels (a and b series) illustrate the head expression, emphasizing pharyngeal patterns for both reporters; the arrowhead indicates expression in the posterior pharyngeal bulb. Note that expression of the mCh-tagged wild-type alh-6 transgene is strong in the posterior bulb of the pharynx (b and b”; arrowhead) and completely suppresses the mutant pattern of gst-4::gfp expression in this tissue (a’ and a”; arrowhead). A similar comparison of mutant and rescue transgene expression is shown for midbody BWMs and hypodermal cells (HYPs) (c and d series), with BWM expression of alh-6::mCh (carets) suppressing gst-4::gfp expression. The arrowhead (d’ and d”) points to a single, nonrescued BWM cell still expressing gst-4::gfp; background gut auto-fluorescence captured in the GFP channel is also visible in these images.
To further confirm that the xrep-2(k1018) and xrep-2(k1022) mutations were loss-of-function alleles of alh-6, we employed two additional approaches. First, we demonstrated that the aberrant gst-4::GFP reporter patterns observed in xrep-2 mutants could be induced following alh-6 RNAi (Figure 1B). Second, we rescued one of the mutant strains (MJCU1018) using either a wild-type alh-6 genomic fragment or a similar genomic construct containing a C-terminal fusion to mCherry (Figure 1C). When introduced into the mutant alh-6(k1018); gst-4::gfp strain, both constructs suppressed the constitutive mutant gst-4::gfp reporter expression pattern in both pharyngeal (Figure 1, A and B) and BWMs (Figure 1, C and D). These findings were consistent with the nearly ubiquitous tissue distribution observed with the mCherry-tagged rescuing transgene. Taken together, we hypothesized that loss of ALH-6 activity results in the accumulation of a proline catabolism intermediate (PC5) that triggers an endogenous toxic signal, activating the phase II detoxification pathway, including the expression of gst-4.
XREP-4 is essential for the phase II stress response to both endogenous and exogenous toxins
A single xrep-4 mutation was isolated as a recessive allele that fails to express the gst-4::gfp reporter in the presence of acrylamide (Figure 2A) (Hasegawa and Miwa 2010). The xrep-4 genetic locus interval was identified by WGS and Hawaiian SNP mapping as described above, and delimited to chromosome I between positions 3–11 Mb; the interval contained nonsynonymous mutations in 26 candidate genes. To determine which of these genes corresponded to xrep-4, we took advantage of the constitutive gst-4::gfp reporter signal in the muscle tissues of alh-6 mutants. We reasoned that XREP-4 activity might be required for this constitutive expression pattern and confirmed that the xrep-4 mutation prevented constitutive gst-4::gfp expression caused by loss of alh-6 (Figure 2B). By using injection RNAi or feeding RNAi for candidate genes for which injection RNAi resulted in embryonic lethality, 12 of the 26 candidate genes from the mapped interval were screened. Of those tested, only RNAi directed against F46F11.6 in the alh-6(k1018) background blocked the constitutive gst-4::gfp signal in muscle (Figure 2C).
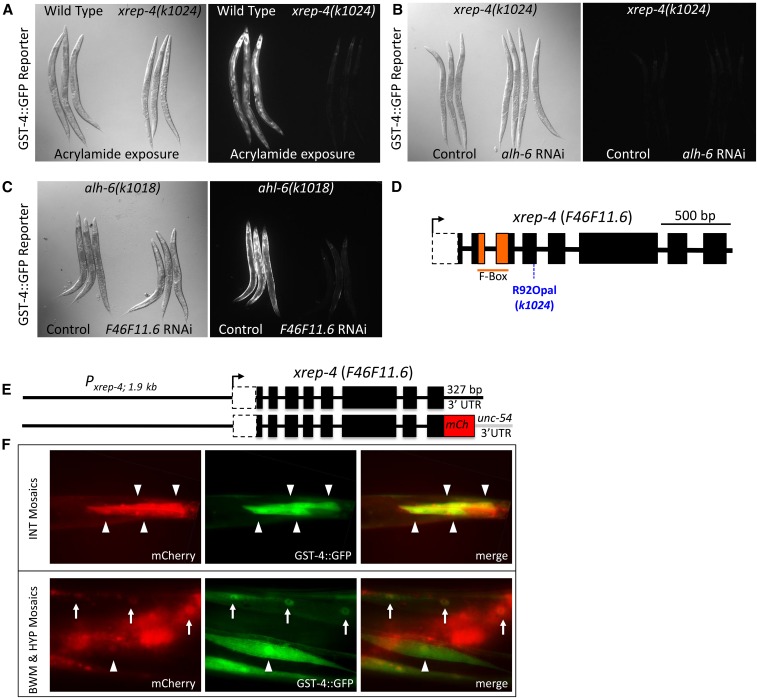
Figure 2.
Phenotypes and the identification of xrep-4 as F46F11.6. (A) The xrep-4 mutants do not induce robust gst-4::gfp reporter expression in response to toxins. A comparison of wild-type and xrep-4(k1024) mutant adult animals harboring the gst-4::gfp translational fusion reporter after exposure to acrylamide for ∼24 hr. Wild-type animals show a robust gst-4::gfp response whereas there is little to no response in xrep-4(k1024) mutants. (B) The xrep-4;gst-4::gfp phenotype is epistatic to alh-6 RNA interference (RNAi). In a wild-type background, alh-6 RNAi is sufficient to induce robust gst-4::gfp expression in bodywall muscles (BWMs) (see Figure 1B). However, little to no gst-4::gfp is detected following alh-6 RNAi in the xrep-4(k1024) mutant background. (C) Phenocopy of the xrep-4 mutation by F46F11.6 RNAi. The alh-6(k1018) mutant results in constitutive gst-4::gfp expression, a phenotype that was exploited to test candidate genes from the xrep-4 mapped interval for their ability to phenocopy xrep-4(k1024). Knockdown of F46F11.6 alone among tested genes was sufficient to block the constitutive alh-6(k1018); gst-4::gfp reporter expression, phenocopying the xrep-4(k1024) mutants. (D) xrep-4 mutants have a premature stop codon mutation in F46F11.6. A single mutation in the F-box-encoding gene F46F11.6 was identified in xrep-4(k1024) mutant animals corresponding to an Arg92 to Opal92 stop codon in the fourth exon. (E) Rescue of xrep-4 mutants with F46F11.6 genomic constructs. Genomic wild-type and mCherry (mCh)-tagged F46F11.6-rescuing constructs are diagramed, each of which was introduced separately to xrep-4(k1024) mutant animals harboring the gst-4::gfp reporter gene. Both genomic clones rescued the xrep-4 mutant phenotype. (F) xrep-4 activity is tissue- and cell-specific. Mosaic expression of a nonintegrated mCh-tagged xrep-4 genomic clone is shown relative to the gst-4::gfp reporter in transgenic xrep-4(k1024) mutant animals. Intestinal (INT) cell expression is highlighted in the top panels with arrowheads pointing to individual cells. The bottom panels highlight BWM (arrowheads) and HYP (arrows) expression patterns in these mosaic transgenic rescue strains. Note that in all cases, the gst-4::gfp signal is present only in cells that are also xrep-4::mCh-positive, demonstrating that transgenic xrep-4 expression driven by the endogenous promoter is sufficient to activate this stress response reporter gene, even in the absence of an exogenous stressor.
WGS of the xrep-4(k1024) mutant strain (MJCU1024) identified a single mutation in F46F11.6 resulting in a premature termination signal, due to an Arg to Opal stop codon substitution (Figure 2D); the predicted translational product lacks the C-terminal two-thirds of the protein. To validate that loss of F46F11.6 activity represents xrep-4, we assayed mutant rescue with a wild-type F46F11.6 genomic fragment or a similar construct containing a C-terminal fusion to mCherry (Figure 2E). Both transgenes restored the ability of xrep-4 mutant animals to activate the gst-4::gfp reporter gene even in the absence of additional stressors and transgene mosaicism indicated that this xrep-4 activity was cell autonomous in both intestinal and muscle tissues (Figure 2E); we used RNAi to confirm that the activation of gst-4::gfp in these strains was SKN-1-dependent (data not shown). We concluded that xrep-4(k1024) is an allele of the F-box-encoding gene F46F11.6 that acts genetically downstream of alh-6. Moreover, XREP-4 mediates the response to both endogenous (presumably P5C) and exogenous (acrylamide) toxic stress in activating the phase II stress response pathway.
XREP-4 functions tissue-autonomously in activating the phase II detoxification response
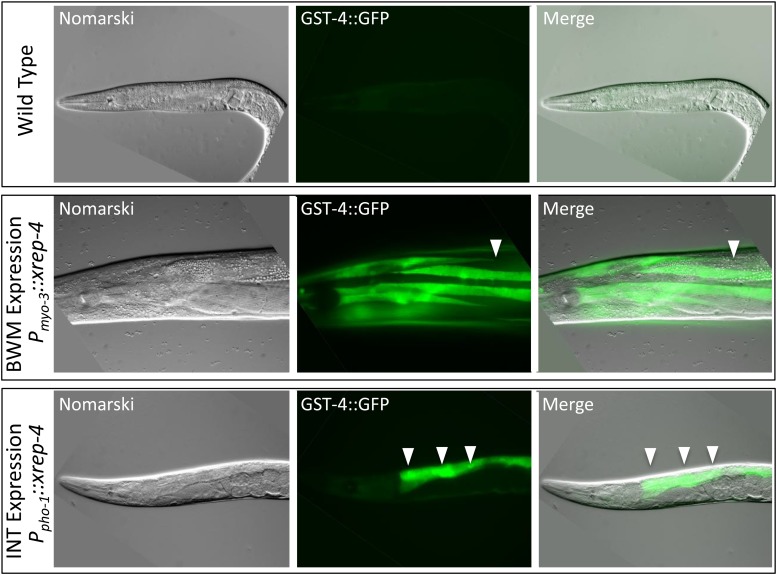
The differential response of our gst-4::gfp reporter in tissues such as muscle and intestine suggested that XREP-4 sensed stress in a tissue-specific manner. To address this directly, we generated constructs in which the coding region of xrep-4 was driven by either a strong muscle (myo-3) or intestinal (pho-1) promoter. BWM expression of the wild-type xrep-4 coding region was able, on its own, to induce the gst-4::gfp reporter specifically in this tissue (Figure 3, middle row). Because the ectopically-expressed xrep-4 transgenes were maintained on a mitotically unstable extrachromosomal array, we were also able to determine (as above) that this activity was cell autonomous. Cell autonomous overexpression of xrep-4 in the intestinal cells was also able to induce the gst-4::gfp reporter (Figure 3, bottom row). Additionally, we noticed that there was variability among resulting transgenic strains in the levels of expression and that the level of transgene expression correlated with strain viability; high levels of xrep-4 activity were not tolerated. We concluded that boosting the levels of XREP-4 above baseline levels was sufficient to trigger a cell- and tissue-specific stress response, suggesting that an acute upregulation of XREP-4 underlies the normal stress response to endogenous and exogenous toxins.
Figure 3.
Tissue-specific expression of xrep-4 is sufficient to induce cell-autonomous gst-4::gfp expression. To address whether XREP-4 activity was cell autonomous, we generated transgenic strains in the gst-4::gfp translational fusion reporter background in which the wild-type xrep-4 coding sequence was driven by strong, tissue-specific promoters, using myo-3 for muscle expression and pho-1 for intestinal expression. In the absence of toxins, the gst-4::gfp reporter is silent (top panels). However, when high levels of xrep-4 are generated in both muscle (middle panels) and intestine (bottom panels), robust activation of the gst-4::gfp reporter is observed. The induction of this reporter gene was cell- and tissue type-specific, as revealed by the mosaic nature of these transgenes; an example is shown in the middle row panels where a gap in GFP signal within a bodywall muscle (BWM) quadrant is highlighted (arrowhead). A similar observation was made in intestinal (INT) cells shown in the bottom row of panels with individual INTs identified with arrowheads. As above, induction of gst-4::gfp expression in these strains occurred in the absence of toxin exposure, revealing that high levels of XREP-4 activity alone could trigger this stress response.
xrep-3(k1023) is a gain-of-function allele of skn-1
The xrep-3 mutant phenotype was previously described as a single dominant allele that exhibited constitutive expression of the gst-4::gfp reporter gene (Hasegawa and Miwa 2010). Previous studies have implicated skn-1 as a positive activator of gst-4::gfp (Hasegawa et al. 2008) and identified skn-1 gain-of-function (gof) alleles as dominant activators of gst-4::gfp (Paek et al. 2012). Because xrep-3(k1023) mapped to the same chromosome as skn-1 (Hasegawa and Miwa 2010) and had a gst-4::gfp expression phenotype that was similar to that of skn-1(gof) mutants (Paek et al. 2012), we considered the possibility that xrep-3 might be a mutation in skn-1. We tested this hypothesis by PCR amplification of the skn-1 genomic region from DNA prepared from xrep-3(k1023) animals and Sanger sequencing of the exons. We found a single missense mutation in xrep-3(k1023) that resulted in an Arg to Cys amino acid substitution in SKN-1 (Figure 4). To validate that this change was the causative mutation in xrep-3(k1023), we amplified the genomic region encoding skn-1 from either wild-type or xrep-3(k1023) mutant animals and introduced them separately into xrep-4(k1018) animals carrying the gst-4::gfp reporter. As expected, the wild-type skn-1 genomic sequences did not activate gst-4::gfp in any progeny derived from the 40 injected hermaphrodites. In contrast, the xrep-3(k1023) mutant skn-1 genomic sequences resulted in constitutive gst-4::gfp reporter gene activation in many F1 animals (27 positive F1s from 25 injected hermaphrodites). These results demonstrated that the k1023 mutant of skn-1 is a gain-of-function (gof) allele that was sufficient to activate gst-4::gfp in a cell autonomous manner. In addition, heterozygous xrep-3(k1023) outcross progeny also constitutively activate the gst-4::gfp reporter demonstrating that this allele is indeed dominant, as previously reported (Hasegawa and Miwa 2010). Finally, we determined that skn-1-specific RNAi in the xrep-3(k1023) mutant background abolished gst-4::gfp reporter gene expression (Figure S2B and Table 2). Taken together, our results demonstrated that xrep-3(k1023) is a dominant, gain-of-function allele of skn-1.
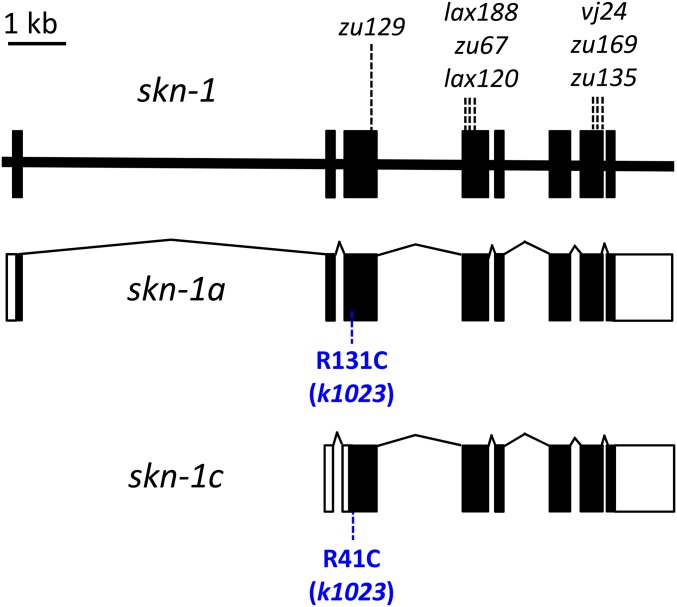
Figure 4.
Identification of xrep-3 as skn-1. The gene structure for the skn-1 locus is diagramed at top with splicing patterns of two transcriptional products (a and c) indicated below. Targeted skn-1 gene sequencing of xrep-3(k1023) mutant genomic DNA identified a single-base change of C to T at position 5,655,485 (WS250). This mutation results in an Arg to Cys amino acid substitution, as shown in blue, corresponding to position 131 or 41 in SKN-1a and SKN-1c isoforms, respectively; additional, previously identified alleles are indicated above the gene structure.
Table 2. Effects of stress pathway component perturbations on gst-4p::gfp expression.
| Genotype | GST-4::GFP levels | |
|---|---|---|
| Untreated | Acrylamide | |
| Wild-type | − | ++ |
| alh-6(k1018) | ++ | +++ |
| xrep-4(k1024) | − | − |
| skr-1/2(RNAi) | − | − |
| wdr-23(k1007) | +++ | N.D. |
| skn-1(k1023)(gof) | +++ | N.D. |
| skn-1(RNAi) | − | − |
| alh-6(k1018); skr-1/2(RNAi) | − | N.D. |
| alh-6(k1018); skn-1(RNAi) | − | N.D. |
| xrep-4(k1024); alh-6(RNAi) | − | N.D. |
| xrep-4(k1024); wdr-23(RNAi) | +++ | N.D. |
| xrep-4(RNAi); skn-1(k1023)(gof) | +++ | N.D. |
| skn-1(k1023)(gof); skn-1(RNAi) | − | − |
N.D., not determined.
XREP-4 genetically functions upstream of both WDR-23 and SKN-1 in regulating the phase II stress response
XREP-4 encodes an F-box protein (Figure 2D), one of 326 such members of this family predicted to be present in C. elegans (Kipreos and Pagano 2000; Dankert et al. 2017). The F-box is a protein–protein interaction motif of ∼50 aa, first identified as components of SCF (Skp1, Cullin, and F-box protein) ubiquitin ligase complexes required for ubiquitin-mediated proteolysis; F-box proteins have since been shown to be required in other cellular processes, including chromosome segregation, transcriptional elongation, and translational control (Kipreos and Pagano 2000; Dankert et al. 2017).
Since xrep-4 remains largely uncharacterized in C. elegans, we sought to place it in the stress response pathway and determine how it might function. Specifically, we were interested in the relationship between XREP-4, the SCF ubiquitin ligase complex, and the stress pathway components WDR-23 (originally identified as XREP-1) (Hasegawa and Miwa 2010) and SKN-1 (identified above as XREP-3). XREP-4 has been reported to physically interact with SKR-1 in high-throughput protein interaction screens (Boxem et al. 2008). SKR-1 and -2, nearly identical proteins, are related to the SCF ubiquitin ligase complex member Skp-1, a known F-box-interacting protein (Nayak et al. 2002; Yamanaka et al. 2002). SKR-1/2 have been linked to the regulation of gst-4 expression via WDR-23 and SKN-1 (Wu et al. 2016), and WDR-23 has been shown to interact with a CUL-4 SCF ubiquitin ligase complex to regulate nuclear SKN-1 levels and activity (Choe et al. 2009).
We were interested in genetically ordering the function of XREP-4 relative to WDR-23, SKN-1, and SKR-1/2 in activating our gst-4::gfp reporter gene in response to both endogenous and exogenous stresses. A complication to exploring these epistatic relationships is that strong loss-of-function mutations in many of these pathway components cause embryonic or early larval lethality or larval arrest. Therefore, we combined genetic mutants in individual factors with RNAi of secondary genes to order known components in the pathway relative to either endogenous and/or exogenous toxins.
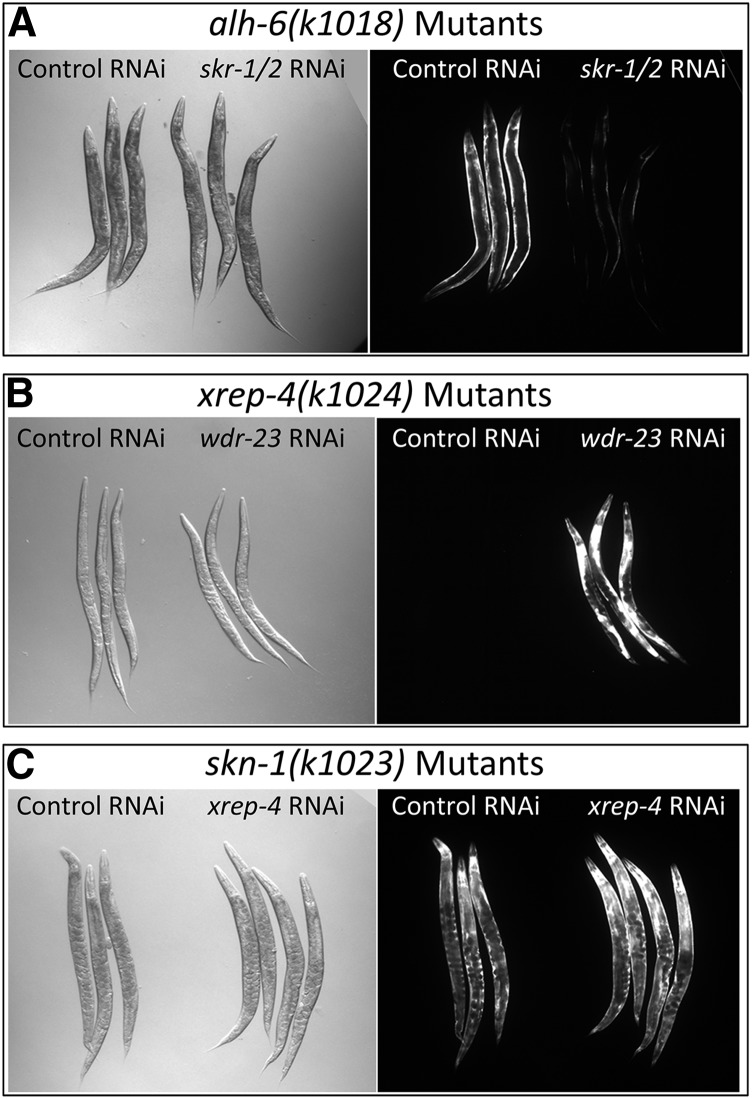
The results of epistasis testing are summarized in Table 2. Expression of gst-4::gfp is induced either by exogenous stress via acrylamide exposure or endogenous stress via loss of ALH-6 activity; in the latter case, the most robust response is seen in muscle tissue. When alh-6(k1018); gst-4::gfp mutant animals are also exposed to acrylamide, gst-4::gfp is further induced in many tissues, including the pharynx, hypodermis, and intestine (Figure S1). We found that the response to endogenous stress in alh-6 mutants was dramatically reduced by RNAi knockdown of either skn-1 (Figure S2) or skr-1/2 (Figure 5A), consistent with previously reported roles for these genes (Pang and Curran 2014; Wu et al. 2016). As indicated above (Figure 2, A and B), xrep-4(k1024) mutants failed to respond to either exogenous acrylamide or endogenous toxins resulting from the loss of ALH-6 activity. In contrast, knockdown of wdr-23 activity in an xrep-4(k1024) mutant strongly activated the gst-4::gfp reporter, even in the absence of acrylamide (Figure 5B). This result demonstrates that much of the phase II signaling pathway remains functional in xrep-4 mutants. Finally, loss of xrep-4 activity had no effect on constitutive gst-4::gfp in the skn-1(gof) mutants (Figure 5C), whereas expression was strongly eliminated by targeting skn-1 itself by RNAi (Figure S2B), confirming a previous report (Paek et al. 2012). Different exogenous toxins have been shown to elicit the phase II detoxification response through distinct pathways (Wu et al. 2016), although all converge on the regulation of SKN-1; our results demonstrate that XREP-4 also functions through SKN-1. Taken together, our findings place XREP-4 at an upstream nodal point that senses and/or triggers the phase II detoxification pathway in response to both endogenous and exogenous toxins, with the primary response limited to either BWM or pharyngeal, hypodermal, and intestinal tissues, respectively.
Figure 5.
Epistatic relationships among stress pathway components. (A) skr-1/2 RNA interference (RNAi) is epistatic to alh-6. The alh-6(k1018) mutants constitutively activate the gst-4::gfp reporter gene that is most obvious in bodywall muscles. Knockdown of the nearly identical Skp-related genes skr-1 and skr-2 by RNAi results in a severe reduction of the gst-4::gfp signal in the alh-6(k1018) mutant background, placing skr-1/2 downstream of alh-6 in the stress response pathway. (B) wdr-23 RNAi is epistatic to xrep-4. The xrep-4(k1024) mutants are unable to induce the gst-4::gfp translational fusion reporter gene expression in response to exogenous toxins such as acrylamide. However, knockdown of wdr-23 by RNAi in the xrep-4(k1024) mutant background strongly induces gst-4::gfp reporter gene expression, placing WDR-23 activity downstream of XREP-4. (C) The skn-1(gof) allele is epistatic to xrep-4. The skn-1(k1023) mutation is a dominant gain-of-function allele that results in constitutively active gst-4::gfp expression, even in the absence of stress. That phenotype is unchanged when skn-1(k1023) mutant animals are exposed to xrep-4 RNAi, placing skn-1 downstream of XREP-4.
XREP-4 regulates the stability of WDR-23
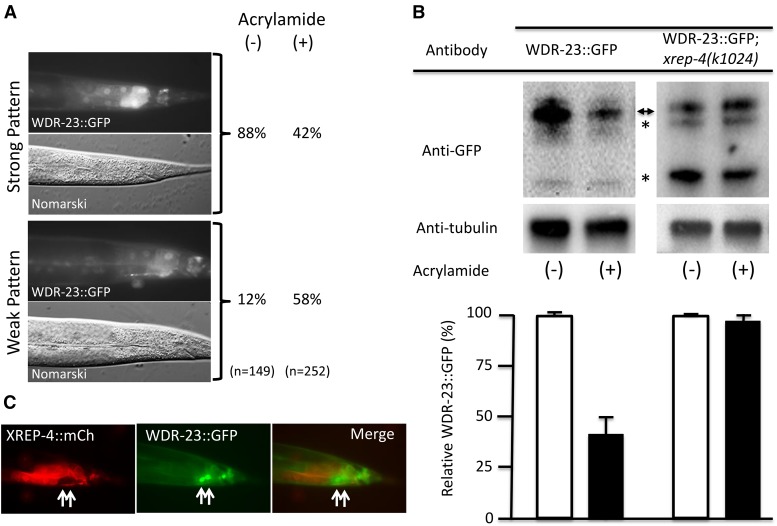
During the course of our studies with the functional genomic WDR-23::GFP translational reporter (Hasegawa and Miwa 2010), we noticed that the levels of WDR-23::GFP in late larval stage animals were dynamic in response to toxins and different mutant or transgenic backgrounds. For example, WDR-23::GFP levels decreased in transgenic animals exposed to acrylamide when compared to unexposed controls (Figure 6A). To quantitate this effect, we compared WDR-23::GFP protein levels by western blotting among age-synchronized (L3 and L4) animals with or without acrylamide exposure (Figure 6B); WDR-23::GFP levels dropped significantly in this population in response to acrylamide. Note that our functional reporter could generate both WDR-23a and WDR-23b isoforms, which are indistinguishable by the anti-GFP antibody used for detection. If XREP-4 was acting as a trigger and/or sensor, then the decrease in WDR-23::GFP due to acrylamide exposure should not occur when XREP-4 activity is lost. Indeed, western analysis shows no change in steady-state levels of WDR-23::GFP after acrylamide exposure in an xrep-4 mutant background (Figure 6B). We also observed reciprocal expression patterns for XREP-4::mCherry and WDR-23::GFP in transgenic animals harboring both functional, translational reporter constructs (Figure 6C). We conclude that XREP-4 functions by reducing the stability of WDR-23 in response to toxins.
Figure 6.
WDR-23 levels are dynamic and dependent on stress and wild-type XREP-4 activity. (A) WDR-23::GFP levels are reduced after acrylamide exposure. Adult animals were scored for WDR-23::GFP signal intensity in posterior intestinal cells after culturing for ∼24 hr in the absence (−) or presence (+) of acrylamide. GFP levels were binned after scoring as either strong (upper panels) or weak (lower panels), revealing that the fraction of animals with strong expression was dramatically reduced (88–42%) after acrylamide exposure. (B) Total WDR-23::GFP protein levels are reduced after acrylamide exposure by an xrep-4-dependent mechanism. Western blots of total protein isolated from L3-L4 stage populations harboring an integrated wdr-23::gfp translational fusion transgene in either a wild-type (left panels) or xrep-4(k1024) mutant background (right panels); these L3–L4 animals had been cultured for ∼24 hr in the absence (−) or presence (+) of acrylamide. After probing with antibodies to detect GFP and the control protein tubulin, all band intensities corresponding to full length WDR-23::GFP (double arrowhead) and presumed degradation products (asterisks) were quantified, normalized to tubulin, and plotted below the corresponding lanes. Marked decreases in the relative GFP levels were detected after acrylamide exposure in the wild-type background. In contrast, WDR-23::GFP levels did not change at all in the xrep-4(k1024) mutant background, although we did note a change in the relative GFP-positive band intensities compared to the wild-type background. (C) XREP-4::mCh and WDR-23::GFP reporter patterns are mutually exclusive. Double transgenic adult animals harboring an extrachromosomal xrep-4::mCh and integrated wdr-23::gfp functional, translational transgene were assayed for reporter gene expression. Intestinal cells with low levels of XREP-4::mCh had strong WDR-23::GFP signals (arrows). Thus, the expression of xrep-4::mCh alone was sufficient to downregulate WDR-23::GFP, even in the absence of acrylamide exposure.
Discussion
C. elegans has emerged as an excellent model to dissect the molecular mechanisms involved in the organismic response to oxidative stress and environmental toxins. The high degree of conservation of disease pathways between C. elegans and higher organisms makes for an effective in vivo genetic model that is amenable to detailed analysis of the responses to such stressors. Toxicology experiments and high-throughput drug screens carried out in C. elegans require a thorough understanding of the detoxification systems in this organism (Hasegawa et al. 2010; Leung et al. 2013; Rangaraju et al. 2015).
In this report, we have molecularly identified several xrep mutants emerging from a genetic screen using a gst-4::gfp translational fusion reporter to measure responses to acrylamide exposure (Hasegawa and Miwa 2010). We employed both whole-genome mapping and candidate gene sequencing strategies to identify the causative mutations, which we confirmed by RNAi phenocopy and transgenic functional assays. Identification of the genes harboring these causative mutations—alh-6 (for xrep-2), the F-box protein encoding F46F11.6 (for xrep-4), and skn-1 (for xrep-3)—has allowed us to define a signaling pathway consistent with the genetic and biochemical properties of these genes (Figure 7, A and B).
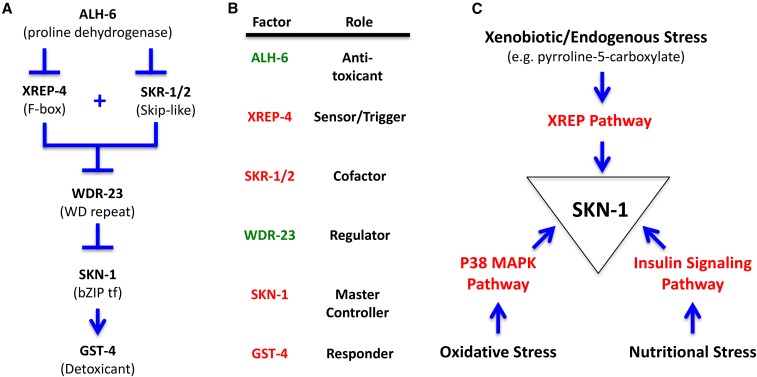
Figure 7.
XREP stress pathway components and relationships. (A) Genetic pathway for the stress response. The stress response pathway based on the genetic results of this study is shown; this pathway is consistent with several previous studies of many of the components (Choe et al. 2009, 2012; Park et al. 2009; Hasegawa and Miwa 2010; Paek et al. 2012; Choe and Leung 2013; Glover-Cutter et al. 2013; Crook-McMahon et al. 2014; Pang et al. 2014; Blackwell et al. 2015; Tang and Pang 2016; Wu et al. 2016). We propose that XREP-4 functions as a key sensor or trigger point in the pathway, the levels of which regulate WDR-23 stability, which in turn regulates SKN-1 transcriptional activity. In nonstress conditions, WDR-23 is able to prevent SKN-1 from activating the pathway (Choe et al. 2009; Hasegawa and Miwa 2010; Tang and Choe 2015; Wu et al. 2016). In our model, reduction or loss of ALH-6 activity results in the buildup of a toxic metabolic intermediate, pyrroline-5-carboxylate, that directly or indirectly upregulates XREP-4 levels. XREP-4 functions with SKR-1/2 to reduce WDR-23 activity in a Skp I, Cul-1, and F-box protein-type ubiquitin-mediated degradation process, releasing SKN-1 that serves as a master transcriptional activator of downstream stress response target genes, including gst-4. (B) Activity relationships and roles among stress pathway components. The stress pathway factors in this study are listed, their role defined, and colored to indicate if their activity promotes suppression (green) or activation (red) of the stress response. (C) Summary of some of the stress pathways operating through SKN-1. Inputs from several stress pathways converging on SKN-1 are shown, including xenobiotic/endogenous stress (this study), oxidative stress through the p38 MAPK Pathway (Wu et al. 2016), and nutritional stress through the insulin signaling pathway [reviewed in Blackwell et al. (2015)].
SKN-1 as a master controller of cellular stress pathways
The C. elegans skn-1 gene encodes an ortholog of the Nrf family of transcription factors that share both Cap‘n’Collar and Basic Region domains (Blackwell et al. 2015). In mammals, these transcription factors regulate many protective and homeostatic pathways including resistance to cytotoxic insults (Blackwell et al. 2015). In contrast to the multiple mammalian Nrf family members, C. elegans has a single skn-1 gene encoding multiple alternatively spliced isoforms that share a core DNA-binding domain. The skn-1 gene was originally identified for its role in early embryonic development (Bowerman et al. 1992), whereas most recent studies have focused on its postembryonic roles in mediating homeostasis and the stress response. Recent work has shown that skn-1 also functions in the unfolded protein response (Choe and Leung 2013), the response to germ cell absence (Steinbaugh et al. 2015), proteasomal regulation (Keith et al. 2016; Lehrbach and Ruvkun 2016; Raynes et al. 2016), and the regulation of autophagy and mitophagy (Salminen and Kaarniranta 2012; Mizunuma et al. 2014; Palikaras et al. 2015a,b,c; Keith et al. 2016).
Intriguingly, many mechanisms that promote C. elegans longevity also increase SKN-1 activity such as the insulin/insulin-like growth factor signaling (IIS) and MAPK pathways (Figure 7C; Blackwell et al. 2015 and references contained therein). Insulin signaling is an important nutrient-dependent mediator of SKN-1 activation and is thought to act through the downstream kinases AKT-1 and AKT-2. SKN-1 is phosphorylated by AKT at multiple positions in vitro and localizes to intestinal nuclei constitutively after mutation of a Ser residue predicted at high stringency to be an AKT target (Blackwell et al. 2015). SKN-1 activity is also typically regulated by signaling through the p38 MAPK pathway. Treatment with oxidative stressors like sodium arsenite activates p38 MAP kinase, and genetic interference with the p38 pathway prevents both SKN-1 nuclear accumulation and impairs resistance to oxidative stress (Blackwell et al. 2015). However, recent evidence suggests that the WDR-23-dependent localization of SKN-1 may be independent of p38 MAPK signaling (Wu et al. 2016).
The SKN-1 transcription factor has previously been implicated as a positive regulator of gst-4 (Hasegawa et al. 2008) and skn-1(gof) alleles as dominant activators of gst-4 (Paek et al. 2012). In our study, xrep-3 was recovered as a dominant constitutive activator of gst-4::gfp (Hasegawa and Miwa 2010). We identified the xrep-3 mutation as an Arg→Cys amino acid substitution in the coding sequence of two splice variants of skn-1, a and c; skn-1 RNAi demonstrated that constitutive gst-4::gfp expression in the xrep-3 strain was dependent upon SKN-1 itself. Our results are consistent with and reinforce the notion proposed by others that SKN-1 is a master controller of the stress pathway (Paek et al. 2012; Blackwell et al. 2015).
WDR-23 regulates SKN-1-dependent expression of gst-4
The initial paper describing the xrep mutants demonstrated that xrep-1 encoded WDR-23, the mammalian homolog of WDR-23 (Hasegawa and Miwa 2010). This protein was originally identified as a WD40 repeat protein that partners with the CUL4/DDB1 ubiquitin ligase complex to regulate the nuclear abundance and transcriptional activity of SKN-1 (Choe et al. 2009). Subsequent work has shown that WDR-23 also plays a role in the regulation of the SKN-1 response to magnesium and pathogens (Papp et al. 2012; Settivari et al. 2013). A growing body of evidence further links the WDR-23/SKN-1 regulatory paradigm to metabolic stress and synaptic function (Papp et al. 2012; Settivari et al. 2013; Staab et al. 2013, 2014). Our genetic analysis exploited the knowledge obtained from an analysis of wdr-23 to define the epistatic relationships with other xrep mutants we have identified molecularly, resulting in the pathways diagramed in Figure 7.
xrep-2 is alh-6, encoding an aldehyde dehydrogenase
In this study, we identified two different alleles of alh-6 that were responsible for the constitutive expression of the gst-4::gfp and gst-30::gfp reporter genes. The alh-6 gene has been previously shown to render worms hypersensitive to ethanol intoxication (Alaimo et al. 2012). In addition, a mutation of alh-6 was shown to accelerate fat mobilization by enhancing fatty acid oxidation and thus reducing survival in response to fasting (Pang et al. 2014). This response, while distinct from the response to toxicants revealed in the current study, was mediated by skn-1. In addition, alh-6 mutants age prematurely when fed Escherichia coli strain OP50 but not HT115 (Pang and Curran 2014), suggesting that alh-6 is linked to monitoring cellular nutrient status and serving a protective role. The constitutive activation of gst-4::gfp we observed in the alh-6 mutants can be reduced by supplementation with glucose (our unpublished data), suggesting that the alh-6 loss-of-function mutants may be triggering a response to both nutrient availability and oxidative stress.
The behavior of both alh-6 mutant alleles as activators of phase II detoxification enzymes (gst-4::gfp and gst-30::gfp) is consistent with a role of alh-6 in degrading P5C (see Discussion below). Whether the amino acid metabolite P5C directly or indirectly triggers the observed detoxification response is currently unknown. The strong activation of GST reporters in muscle tissue may reflect the elevated mitochondrial function, amino acid synthesis and utilization, and protein turnover in the metabolically active tissue.
The mammalian homolog most similar to alh-6 is the NAD-dependent pyrroline-5-carboxylate dehydrogenase gene ALDH4A1. This enzyme catalyzes the irreversible conversion of P5C, derived either from proline or ornithine, to glutamate. In turn, glutamate is a precursor to α-ketoglutarate, the metabolic entry point into the tricarboxylic acid cycle. Mutations in ALDH4A1 that affect enzyme function lead to a human disorder called hyperprolinemia type II, a defect in proline catabolism associated with childhood seizures (Flynn et al. 1989). Our analysis reveals that the two mutations in alh-6 corresponding to the xrep-2 alleles are in a highly-conserved region of the C-terminus. Previously identified mutants of alh-6 also map near this conserved region (see Figure 1A). Alignments of ALH-6 with mouse and human ALDH4A1 reveal remarkable similarity in this region. From the crystal structures of mouse and human ALDH4A1, we infer that the defects in ALH-6 are adjacent to a conserved motif important for interacting with the product glutamate and the cofactor NAD.
xrep-4 is the F-box protein-encoding gene F46F11.6
xrep-4 mutants fail to express gst-4::gfp in the presence of acrylamide. We identified xrep-4 as an allele of the F-box protein-encoding gene F46F11.6, which we validated by transgenic rescue. RNAi of alh-6 in the xrep-4 strain failed to induce gst-4::gfp expression, indicating that xrep-4 acts after alh-6 in the pathway. However, RNAi of wdr-23 in the xrep-4 mutant background led to gst-4::gfp expression. Interestingly, xrep-4 was previously recovered in a genome-wide RNAi screen to identify RNAi clones that reduced intestinal expression of the phase II enzyme gcs-1p::gfp in a prdx-2 (peroxidase) mutant background (Crook-McMahon et al. 2014). However, in that screen, xrep-4 RNAi also decreased the expression of a non-phase II enzyme suggesting a broader role in gene expression (Crook-McMahon et al. 2014).
XREP-4 has been shown to interact with SKR-1, a protein encoded by the skr-1 gene and known partner of F-box proteins that act as regulators of ubiquitination/protein degradation (Boxem et al. 2008). A genome-wide RNAi screen to identify novel regulators that are required for activation of gst-4 during exposure to the electrophile juglone identified skr-1/2 as the only members of this multigene family that were required in this assay (Wu et al. 2016). Based on these observations, we carried out RNAi inactivation of skr-1/2 and found that, like xrep-4 inactivation, gst-4::gfp expression was blocked by skr-1/2 depletion in the alh-6 endogenous stress mutant background.
Properties of XREP-4 and the relationship to WDR-23/SKN-1 regulation
The F-box protein XREP-4 is part of a family of ∼326 F-box proteins in C. elegans. XREP-4 is conserved throughout nematodes, although the F-box domain is the only shared feature. The F-box domain is a motif of ∼50 aa that normally mediates protein–protein interactions. It was first identified in cyclin F and, in this context, the F-box motif interacts directly with the SCF protein SKP1 (Bai et al. 1996). SCF complexes bind to their substrates and target them for ubiquitin-mediated degradation. Our studies are consistent with a role for the F-box protein XREP-4 acting in combination with SKR-1/2 to alter the stability of the downstream target WDR-23. Our RNAi results suggest that both SKR-1/2 and XREP-4 act upstream of WDR-23. It is not known whether SKR-1/2 and XREP-4 act as part of a common CUL-1-based E3 ubiquitin ligase complex or in a parallel pathway. However, our western and in vivo results suggest an antagonistic relationship between XREP-4 and WDR-23::GFP levels, strongly suggesting that the upregulation of XREP-4 in response to stress results in WDR-23 degradation.
Ordering the steps in the pathway of phase II detoxification based on epistasis of the xrep mutants
Our data suggest that the alh-6 (xrep-2) mutation induces endogenous metabolic stress, functioning upstream of the other xrep mutants (Figure 7). The alh-6 mutants constitutively express both gst-4::gfp and gst-30::gfp, consistent with a continuous activation of the cellular detoxification response, likely in response to an accumulation of a toxic proline metabolic intermediate. A key sensor of this toxic stress is the F-box protein-encoding gene xrep-4. XREP-4 functions genetically to block the ability of WDR-23 to inhibit SKN-1 activity, resulting in SKN-1-mediated activation of gst-4 and other detoxification genes. XREP-4 physically interacts with SKR-1 and inactivation of either xrep-4 or skr-1/2 leads to a disruption of the gst-4 induction in response to either acrylamide or loss of alh-6 (Wu et al. 2016). Thus, our genetic evidence suggested that the newly identified XREP-4 F-box protein may interact with SKR-1/2 to influence the stability of WDR-23. We confirmed this effect using a WDR-23::GFP reporter; in response to acrylamide, WDR-23 levels dropped dramatically in wild-type animals, but remained unaltered in the xrep-4 mutants. These epistatic relationships suggest a cascade of inhibitory events in which XREP-4 participates in a selective targeting of WDR-23 to reduce its levels in response to acrylamide. The reduced stability of WDR-23 influences its ability to regulate the activity and localization of SKN-1, which in turn regulates downstream target genes represented by the reporter constructs gst-4::gfp and gst-30::gfp. Thus, the XREP pathway is one of the key regulators of SKN-1 signaling, consorting with insulin signaling and p38 MAPK signaling in mediating the response to various forms of endogenous and exogenous stress (Figure 7C).
Conclusions
One of the key organismic responses to oxidative stress is the transcriptional induction of genes encoding enzymes, such as GST, that serve to eliminate the offending metabolite. In this report, we have characterized several components of a genetic pathway that further defines how a defect in proline catabolism (alh-6) or exogenous stressors such as acrylamide may induce the phase II detoxification system in C. elegans. The cascade of regulatory events triggered by endogenous or exogenous stress is sensed in part by induction of XREP-4, an F-box protein that alters the stability of WDR-23. WDR-23 is a known negative regulator of SKN-1 nuclear entry and transcriptional activation. The xrep pathway leading to the induction of gst-4 and other phase II detoxification enzymes represents an important response to environmental and metabolic oxidative stress. An understanding of the pathways by which toxicants are recognized and eliminated by C. elegans may provide clues as to how this evolutionarily conserved process might be regulated.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.202515/-/DC1.
Acknowledgments
We thank Koichi Hasegawa and Yu Nonomura for xrep mutant strains and information. Our work was facilitated by the WormBase resource. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD-010440). This work was supported, in part, by the Intramural Research Program of the NIH and the National Institute of Diabetes and Digestive and Kidney Diseases.
Note added in proof: During the course of this study, the Choe Lab (Wu et al., 2017) independently isolated multiple alleles of F46F11.6 (xrep-4) in a screen for genes required for the oxidative stress response. Their results support the same relationships between XREP-4, SKR-1, WDR-23, and SKN-1 as those described in this study.
Footnotes
Communicating editor: B. Goldstein
Literature Cited
- Ahringer, J., 2006 Reverse genetics (April 6, 2006), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.47.1, http://www.wormbook.org.
- Alaimo J. T., Davis S. J., Song S. S., Burnette C. R., Grotewiel M., et al. , 2012. Ethanol metabolism and osmolarity modify behavioral responses to ethanol in C. elegans. Alcohol. Clin. Exp. Res. 36: 1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C., Sen P., Hofmann K., Ma L., Goebl M., et al. , 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Steinbaugh M. J., Hourihan J. M., Ewald C. Y., Isik M., 2015. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 88: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B., Eaton B. A., Priess J. R., 1992. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68: 1061–1075. [DOI] [PubMed] [Google Scholar]
- Boxem M., Maliga Z., Klitgord N., Li N., Lemmens I., et al. , 2008. A protein domain-based interactome network for C. elegans early embryogenesis. Cell 134: 534–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell, B., 2015 BBMap short-read aligner, and other bioinformatics tools. Available at: http://sourceforge.net/projects/bbmap/. Accessed July 10, 2015.
- Carroll A. S., Gilbert D. E., Liu X., Cheung J. W., Michnowicz J. E., et al. , 1997. SKN-1 domain folding and basic region monomer stabilization upon DNA binding. Genes Dev. 11: 2227–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe K. P., Leung C. K., 2013. SKN-1/Nrf, a new unfolded protein response factor. PLoS Genet. 9: e1003827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe K. P., Przybysz A. J., Strange K., 2009. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol. Cell. Biol. 29: 2704–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe K. P., Leung C. K., Miyamoto M. M., 2012. Unique structure and regulation of the nematode detoxification gene regulator, SKN-1: implications to understanding and controlling drug resistance. Drug Metab. Rev. 44: 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook-McMahon H. M., Oláhová M., Button E. L., Winter J. J., Veal E. A., 2014. Genome-wide screening identifies new genes required for stress-induced phase 2 detoxification gene expression in animals. BMC Biol. 12: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankert J. F., Pagan J. K., Starostina N. G., Kipreos E. T., Pagano M., 2017. FEM1 proteins are ancient regulators of SLBP degradation. Cell Cycle 16: 556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M., Poole R. J., Sarin S., Bigelow H., Hobert O., 2010. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS One 5: e15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn M. P., Martin M. C., Moore P. T., Stafford J. A., Fleming G. A., et al. , 1989. Type II hyperprolinaemia in a pedigree of Irish travellers (nomads). Arch. Dis. Child. 64: 1699–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison, E., and G. Marth, 2012 Haplotype-based variant detection from short-read sequencing. arXiv Available at: https://arxiv.org/abs/1207.3907.
- Glover-Cutter K. M., Lin S., Blackwell T. K., 2013. Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet. 9: e1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Miwa J., 2010. Genetic and cellular characterization of Caenorhabditis elegans mutants abnormal in the regulation of many phase II enzymes. PLoS One 5: e11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Miwa S., Tsutsumiuchi K., Taniguchi H., Miwa J., 2004. Extremely low dose of acrylamide decreases lifespan in Caenorhabditis elegans. Toxicol. Lett. 152: 183–189. [DOI] [PubMed] [Google Scholar]
- Hasegawa K., Miwa S., Tajima T., Tsutsumiuchi K., Taniguchi H., et al. , 2007. A rapid and inexpensive method to screen for common foods that reduce the action of acrylamide, a harmful substance in food. Toxicol. Lett. 175: 82–88. [DOI] [PubMed] [Google Scholar]
- Hasegawa K., Miwa S., Isomura K., Tsutsumiuchi K., Taniguchi H., et al. , 2008. Acrylamide-responsive genes in the nematode Caenorhabditis elegans. Toxicol. Sci. 101: 215–225. [DOI] [PubMed] [Google Scholar]
- Hasegawa K., Miwa S., Tsutsumiuchi K., Miwa J., 2010. Allyl isothiocyanate that induces GST and UGT expression confers oxidative stress resistance on C. elegans, as demonstrated by nematode biosensor. PLoS One 5: e9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Doniach T., 1997. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics 146: 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Ishii T., Wakabayashi N., Yamamoto M., 1999. Regulatory mechanisms of cellular response to oxidative stress. Free Radic. Res. 31: 319–324. [DOI] [PubMed] [Google Scholar]
- Jakoby W. B., Ziegler D. M., 1990. The enzymes of detoxication. J. Biol. Chem. 265: 20715–20718. [PubMed] [Google Scholar]
- Jones L. M., Rayson S. J., Flemming A. J., Urwin P. E., 2013. Adaptive and specialised transcriptional responses to xenobiotic stress in Caenorhabditis elegans are regulated by nuclear hormone receptors. PLoS One 8: e69956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn N. W., Rea S. L., Moyle S., Kell A., Johnson T. E., 2008. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem. J. 409: 205–213. [DOI] [PubMed] [Google Scholar]
- Keith S. A., Maddux S. K., Zhong Y., Chinchankar M. N., Ferguson A. A., et al. , 2016. Graded proteasome dysfunction in Caenorhabditis elegans activates an adaptive response involving the conserved SKN-1 and ELT-2 transcription factors and the autophagy-lysosome pathway. PLoS Genet. 12: e1005823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos E. T., Pagano M., 2000. The F-box protein family. Genome Biol. 1: REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A., Ohta T., Yamamoto M., 2004. Unique function of the Nrf2-Keap1 pathway in the inducible expression of antioxidant and detoxifying enzymes. Methods Enzymol. 378: 273–286. [DOI] [PubMed] [Google Scholar]
- Lehrbach N. J., Ruvkun G., 2016. Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1. Elife 5: e17721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C. K., Wang Y., Malany S., Deonarine A., Nguyen K., et al. , 2013. An ultra high-throughput, whole-animal screen for small molecule modulators of a specific genetic pathway in Caenorhabditis elegans. PLoS One 8: e62166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Matilainen O., Jin C., Glover-Cutter K. M., Holmberg C. I., et al. , 2011. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 7: e1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsubuchi H., Nakamura K., Matsumoto S., Endo F., 2008. Inborn errors of proline metabolism. J. Nutr. 138: 2016S–2020S. [DOI] [PubMed] [Google Scholar]
- Mizunuma M., Neumann-Haefelin E., Moroz N., Li Y., Blackwell T. K., 2014. mTORC2-SGK-1 acts in two environmentally responsive pathways with opposing effects on longevity. Aging Cell 13: 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S., Santiago F. E., Jin H., Lin D., Schedl T., et al. , 2002. The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 12: 277–287. [DOI] [PubMed] [Google Scholar]
- Nguyen T., Nioi P., Pickett C. B., 2009. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 284: 13291–13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osburn W. O., Kensler T. W., 2008. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutat. Res. 659: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek J., Lo J. Y., Narasimhan S. D., Nguyen T. N., Glover-Cutter K., et al. , 2012. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab. 16: 526–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Lo M. C., Schmidt D., Pelczer I., Thurber S., et al. , 1997. Skn-1: evidence for a bipartite recognition helix in DNA binding. Proc. Natl. Acad. Sci. USA 94: 5556–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K., Lionaki E., Tavernarakis N., 2015a Coupling mitogenesis and mitophagy for longevity. Autophagy 11: 1428–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K., Lionaki E., Tavernarakis N., 2015b Interfacing mitochondrial biogenesis and elimination to enhance host pathogen defense and longevity. Worm 4: e1071763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K., Lionaki E., Tavernarakis N., 2015c Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521: 525–528. [DOI] [PubMed] [Google Scholar]
- Pang S., Curran S. P., 2014. Adaptive capacity to bacterial diet modulates aging in C. elegans. Cell Metab. 19: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S., Lynn D. A., Lo J. Y., Paek J., Curran S. P., 2014. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat. Commun. 5: 5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp D., Csermely P., Sőti C., 2012. A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans. PLoS Pathog. 8: e1002673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. K., Tedesco P. M., Johnson T. E., 2009. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell 8: 258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybysz A. J., Choe K. P., Roberts L. J., Strange K., 2009. Increased age reduces DAF-16 and SKN-1 signaling and the hormetic response of Caenorhabditis elegans to the xenobiotic juglone. Mech. Ageing Dev. 130: 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju S., Solis G. M., Petrascheck M., 2015. High-throughput small-molecule screening in Caenorhabditis elegans. Methods Mol. Biol. 1263: 139–155. [DOI] [PubMed] [Google Scholar]
- Raynes R., Juarez C., Pomatto L. C., Sieburth D., Davies K. J., 2016. Aging and SKN-1-dependent loss of 20S proteasome adaptation to oxidative stress in C. elegans. J. Gerontol. A Biol. Sci. Med. Sci. 72: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2016. R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Roh J. Y., Park Y. J., Choi J., 2009. A cadmium toxicity assay using stress responsive Caenorhabditis elegans mutant strains. Environ. Toxicol. Pharmacol. 28: 409–413. [DOI] [PubMed] [Google Scholar]
- Rupert P. B., Daughdrill G. W., Bowerman B., Matthews B. W., 1998. A new DNA-binding motif in the Skn-1 binding domain-DNA complex. Nat. Struct. Biol. 5: 484–491. [DOI] [PubMed] [Google Scholar]
- Salminen A., Kaarniranta K., 2012. AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res. Rev. 11: 230–241. [DOI] [PubMed] [Google Scholar]
- Schlipalius D. I., Valmas N., Tuck A. G., Jagadeesan R., Ma L., et al. , 2012. A core metabolic enzyme mediates resistance to phosphine gas. Science 338: 807–810. [DOI] [PubMed] [Google Scholar]
- Settivari R., Vanduyn N., Levora J., Nass R., 2013. The Nrf2/SKN-1-dependent glutathione S-transferase π homologue GST-1 inhibits dopamine neuron degeneration in a Caenorhabditis elegans model of manganism. Neurotoxicology 38: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D., Singh R. K., Moxley M. A., Henzl M. T., Becker D. F., et al. , 2012. The three-dimensional structural basis of type II hyperprolinemia. J. Mol. Biol. 420: 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab T. A., Griffen T. C., Corcoran C., Evgrafov O., Knowles J. A., et al. , 2013. The conserved SKN-1/Nrf2 stress response pathway regulates synaptic function in Caenorhabditis elegans. PLoS Genet. 9: e1003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab T. A., Evgrafov O., Egrafov O., Knowles J. A., Sieburth D., 2014. Regulation of synaptic nlg-1/neuroligin abundance by the skn-1/Nrf stress response pathway protects against oxidative stress. PLoS Genet. 10: e1004100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbaugh M. J., Narasimhan S. D., Robida-Stubbs S., Moronetti Mazzeo L. E., Dreyfuss J. M., et al. , 2015. Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. Elife 4: e07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis G. P., Bohmann D., 2010. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci. Signal. 3: re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Pang S., 2016. Proline catabolism modulates innate immunity in Caenorhabditis elegans. Cell Rep. 17: 2837–2844. [DOI] [PubMed] [Google Scholar]
- Tang L., Choe K. P., 2015. Characterization of skn-1/wdr-23 phenotypes in Caenorhabditis elegans; pleiotrophy, aging, glutathione, and interactions with other longevity pathways. Mech. Ageing Dev. 149: 88–98. [DOI] [PubMed] [Google Scholar]
- Wang J., Robida-Stubbs S., Tullet J. M., Rual J. F., Vidal M., et al. , 2010. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 6: e1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H., 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. W., Deonarine A., Przybysz A., Strange K., Choe K. P., 2016. The Skp1 homologs SKR-1/2 are required for the Caenorhabditis elegans SKN-1 antioxidant/detoxification response independently of p38 MAPK. PLoS Genet. 12: e1006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. W., Wang Y., Choe K. P., 2017. F-box protein XREP-4 is a new regulator of the oxidative stress response in Caenorhabditis elegans. Genetics 206: 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka A., Yada M., Imaki H., Koga M., Ohshima Y., et al. , 2002. Multiple Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with Cullins and F-box proteins. Curr. Biol. 12: 267–275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and genomic sequences are available upon request. All oligonucleotides used for cloning are listed in Table S2.