Figure 7. RNA editing slows down cephalopod genome evolution.
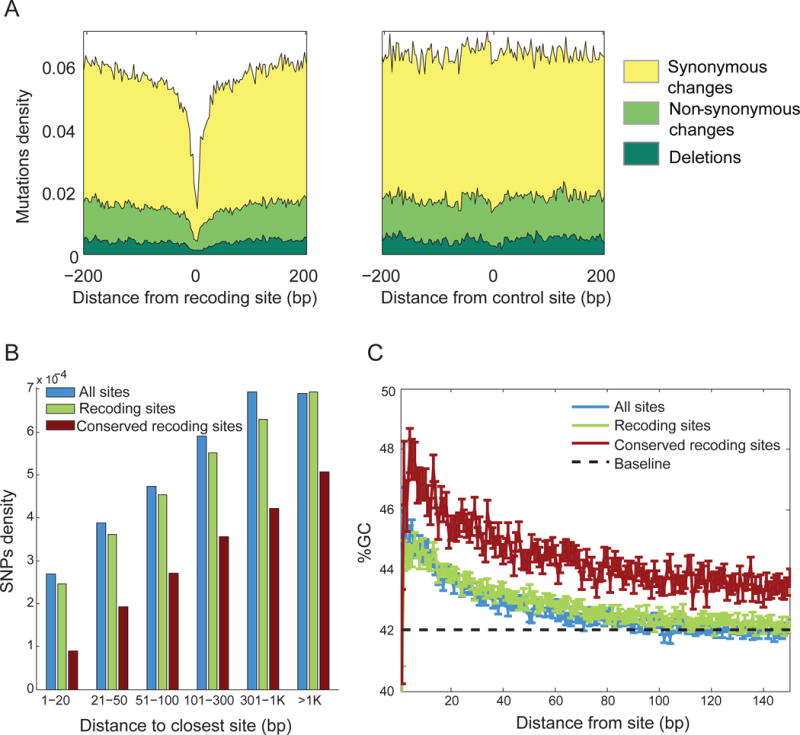
(A) Inter-species mutations are purified from genome loci surrounding conserved recoding sites (data shown for sites shared by squid and sepia). Depletion of mutations extends up to ~100bp of shared recoding sites (left). As a control, we show the mutations density (mutations/bp) around random non-edited adenosines from the same transcripts (right). Yellow – synonymous change; light green – non-synonymous; dark green – deletions.
(B) Genomic polymorphisms are depleted near editing/recoding/conserved-recoding sites in squid, attesting to reduced genome plasticity. Effect is stronger for recoding sites, and even more so for the conserved recoding sites.
(C) GC-content is elevated near editing sites in squid, allowing for more stable double-stranded RNA structures. The effect is even stronger in conserved sites. Dashed line represents the baseline GC level in the entire ORFome, and error bars represent the S.E.M. See Supp. Fig. 3 for analyses similar to those presented in panels A–C in other species.
