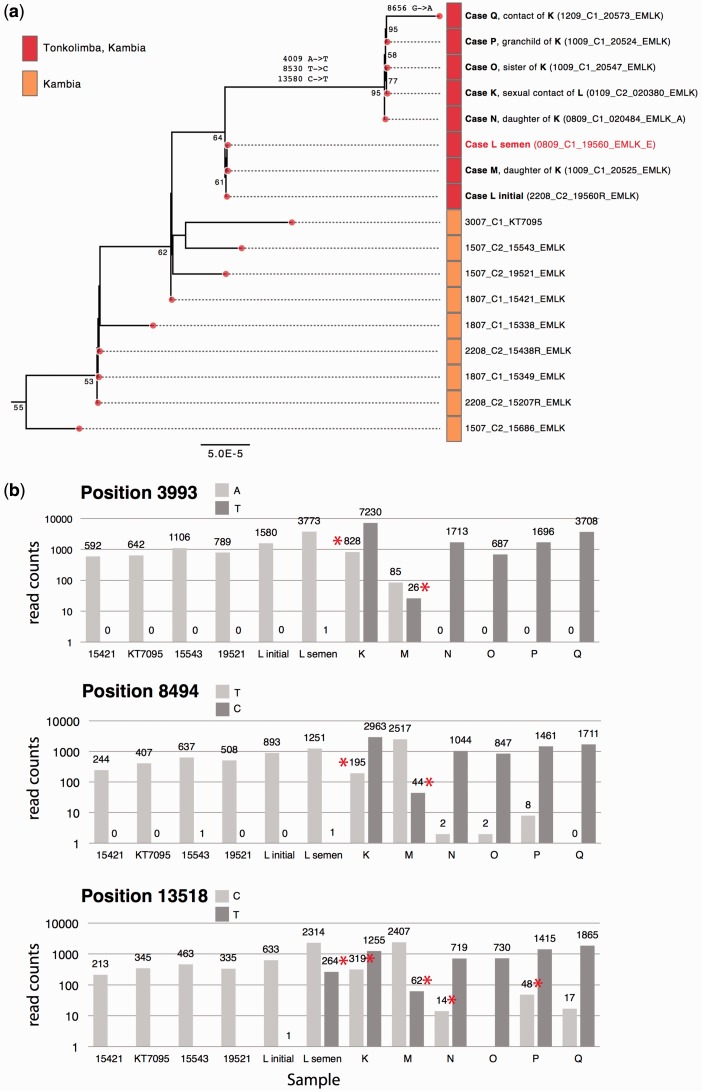
Figure 5.
(a) Maximum-likelihood tree showing the Kambia cluster with possible sexual transmission and full genome from a semen sample. The virus from case L’s initial acute sample (19560R_EMLK, GenBank accession no. KU296580) is the most probable index case of the Kambian cluster. After a 21-d period of quarantine, case L was discharged on 18 July 2015. A sample from case L’s semen (19560_EMLK, labeled in red, GenBank accession no. KU296821) was collected on 7 September 2015. The virus genome isolated from the deceased case K (020380_EMLK, GenBank accession no. KU296775) is genetically identical to case L, which also clusters closely with case M (K’s 23-year-old daughter, 20525_EMLK, GenBank accession no. KU296487). For each cluster case, minority variants for three key positions can be found in (b). Symptom onset of case M (3 September 2015) was 15 d later than onset of case K (26 August 2015). Case K is genetically identical to three known contacts of K: case N (020484_EMLK, older daughter of K, GenBank accession no. KU296462), case O (20547_EMLK, sister of K, GenBank accession no. KU296455), case P (20524_EMLK, grandchild of K, GenBank accession no. KU296424). Case Q (20573_EMLK, GenBank accession no. KU296654), also from the same village, is the most recent sampled case from this cluster. The lineage is related to earlier viruses from lineage F (19521_EMLK, 15543_EMLK, KT7095, and 15421_EMLK, see Fig. 2). See legend of Fig. 3(b) for additional figure details. (b) Minor variants in the Kambia lineage. In genomes from the Kambia cluster (a) three genome positions (3993, 8494, and 13518) showed changes across the entire lineage leading from 19521 through to all genomes in the family cluster. The presence of each of the two variant nucleotides was counted in the raw read set for each sample to gain additional information about possible transmission patterns. Positions with minor variants at >1% frequency are marked with a red asterisk. Positions 3994, 8496, and 13520 showed mixed nucleotides in samples from cases K and M, similar to the case L semen sample (but not in the case L initial sample). Later cases in the lineage (N–Q) showed predominately one of the variants at each of the three positions, although position 13520 showed some persistence of the minor variant C. These data further support the phylogenetic conclusions based on the consensus genome sequence with the L semen sample containing minor variants at the three positions that increase in frequency in samples from cases K and M and become the dominant nucleotide in cases N–Q.