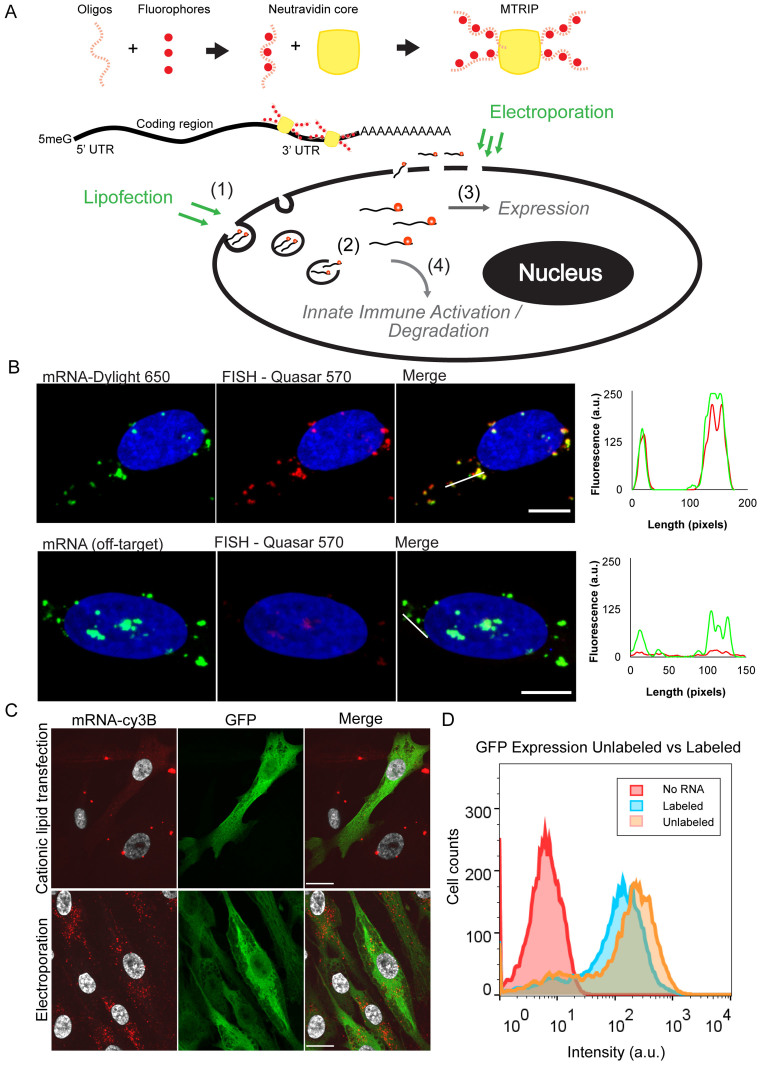
Figure 1.
mRNA labeling, validation, and transfection into cells using cationic lipids or electroporation. (A) Illustrative diagram of mRNA labeling and delivery. MTRIPS are composed of four biotinylated and fluorescently labeled oligonucleotides assembled on a Neutravidin core. They bind to IVT mRNA in the 3΄ UTR region. Fluorescently labeled mRNA is electroporated or lipofected into cells. Electroporated mRNA is immediately available in the cytosol, while lipofected mRNA must first travel through endosomes until released. Degradation and sequestration of mRNA reduces final expression of the target protein. Highlighted mechanisms of importance include (i) mRNA entry pathway, (ii) cytoplasmic release, (iii) translational efficiency (RNA/protein correlation) and (iv) innate immune (PKR) activation. (B) Labeled mRNA (green) delivered via lipofection colocalized with Stellaris FISH probe signal (red). Line profiles are indicated by white lines. No FISH signal was visible in cells transfected with an mRNA containing a different coding region. (scale bar = 10 μm). (C) Cationic lipid transfection and electroporation with labeled mRNA encoding EGFP resulted in similar levels of protein production (green) but distinct subcellular distributions of mRNA (red) at 2 h post-transfection. Upon electroporation, small mRNA granules were observed throughout the cell cytoplasm, while cationic lipid transfection resulted in few large mRNA granules (scale bar = 20 μm). (D) Flow cytometry of EGFP expression using labeled versus unlabeled EGFP mRNA showed only a slight reduction in protein expression.