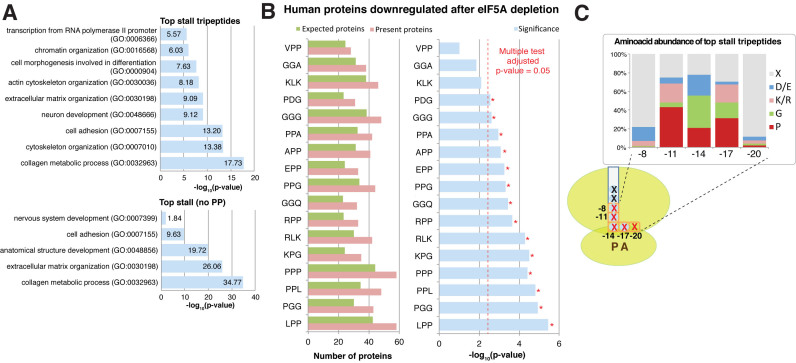
Figure 6.
Conservation of the identified motifs in the human proteome. (A) Selected gene ontology terms significantly enriched in the human proteins with a high content (>25) of eIF5A-dependent motifs, including those tripeptides that contained PP or not. (B) Down-regulated proteins after eIF5A depletion (37) are enriched for the motifs that causer an eIF5A-dependent pause. Randomly expected proteins (green), identified (pink) and significance (according to the hypergeometric distribution, blue). Significance level for Bonferroni-adjusted multiple testing is depicted in red and indicated by *. Only the motifs present in at least 40 proteins identified by Fujimura et al. (37) were considered for the analysis. (C) Amino acid abundance in relation to the ribosome structure for those motifs associated with eIF5A-dependent pausing in S. cerevisiae.