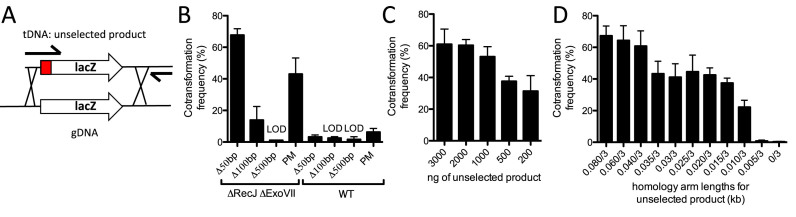
Figure 2.
Efficient cotransformation of ssDNA exonuclease mutants using mutant constructs made in a single PCR reaction. (A) Schematic indicating how 0.08/3 kb unselected products are generated in a single PCR reaction. The mutation in the unselected product is indicated as a grey box and the relative positions of the oligonucleotides used for amplification are highlighted by black arrows. (B) Cotransformation assays using 50 ng of a selected product and 3000 ng of an unselected product (all are 0.08/3 kb) into the indicated strain backgrounds. The different unselected products tested generate the indicated type of mutation in the lacZ gene. (C) Cotransformation assays in a ΔrecJ ΔexoVII mutant using 50 ng of a selected product and the indicated amount of an unselected product (0.08/3 kb) that introduces a 50 bp deletion into the lacZ gene. (D) Cotransformation assays in a ΔrecJ ΔexoVII mutant using 50 ng of a selected product and 3000 ng of an unselected product (X/3 kb) that introduces a 50 bp deletion into the lacZ gene. All data are from at least three independent biological replicates and shown as the mean ± SD. LOD = limit of detection and PM = point mutation.