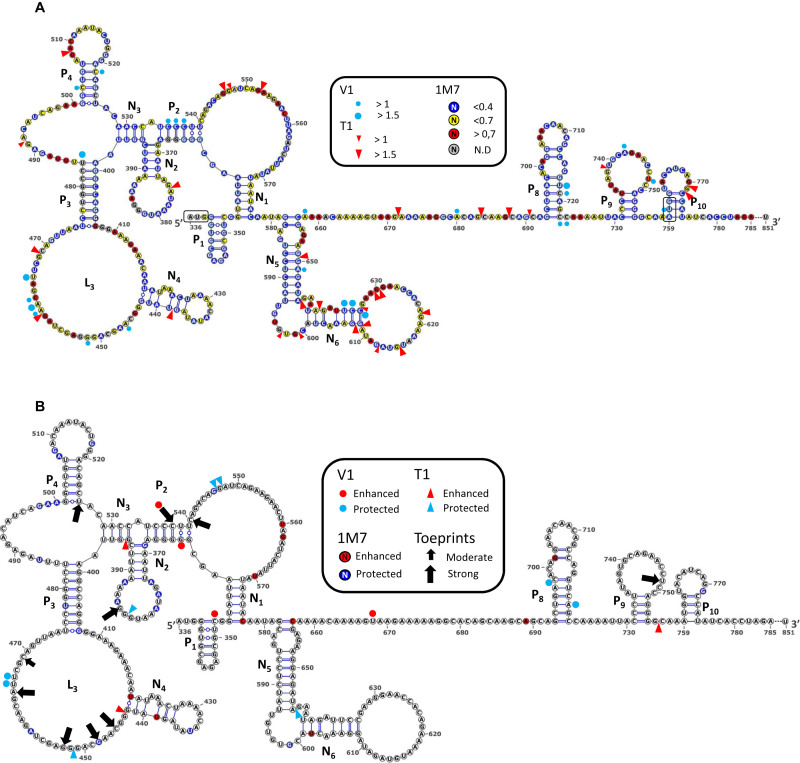
Figure 1.
RNA secondary structure model of the HIV1 Gag-IRES and 40S ribosome subunit footprints and toeprints. (A) Schematic representation of the secondary structure model of Gag-IRES. Nucleotides are colored according to their reactivity toward 1M7 as indicated in the box. Red triangles and blue dots represent RNAse T1 and V1 cleavages. Pairings numbering (Pn and Nn) are as described in the text. Nucleotide numbering is from the +1 of transcription (First nucleotide of TAR). Experimental values result from the mean of three independent experiments (see material and methods and supplementary material). (B) Footprints and toeprints of the 40S ribosomal subunit on Gag-IRES. As indicated in the box, the colors and signs compare the reactivities without and with saturating concentration of 40S ribosomal subunit. Nucleotides in red are more reactive to 1M7, while nucleotides in blue are less. Red and Blue triangles indicate nucleotides exposed or protected to RNAse T1 respectively. Red and Blue dots indicate nucleotides exposed or protected to RNAse V1 respectively. Black Arrows indicate premature RT stops observed in the presence of 40S ribosomal subunits. Small and big arrows are respectively moderate and strong toeprints. Experimental values result from the mean of three independent experiments (see material and methods and supplementary material).