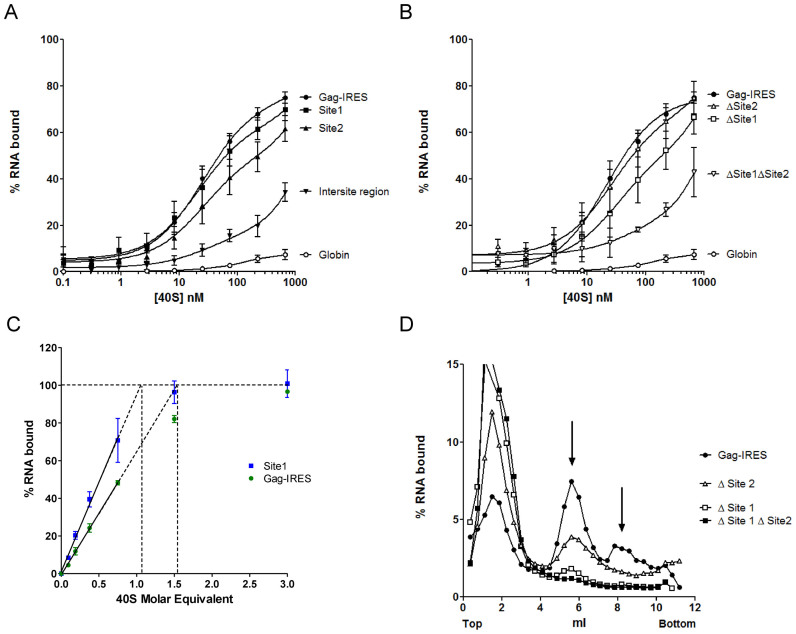
Figure 3.
Two sites within the Gag IRES can independently recruit the 40S ribosomal subunit. (A) Binding curves of 32P-labeled Site 1 (▪, G367–U486), Site 2 (▴, G630–A751), full length HIV-1 Gag-IRES (•, A336–U851), the intersite region (▾), and the globin transcript (○) with purified 40S ribosomal subunits. (B) Binding curves for HIV-1 Gag-IRES deleted of site 1 (Δ Site 1: A336–U851Δ363–485), of site 2 (Δ Site 2: A336-U851Δ632–756), or of both sites (Δ Site 1 Δ Site 2: A336–U851Δ363–485Δ632–756) as compared to the full length HIV-1 Gag-IRES (A336–U851), and to the globin transcript. (C) Determination of the stoichiometry of the 40S ribosomal subunit and the Gag-IRES RNA. A constant concentration of RNA corresponding to the whole IRES (▪) or to the isolated site 1 (G367–U486 •), tenfold over the Kd of the Gag-IRES/40S complex (250 nM) was incubated with increasing concentration of 40S ribosome. The proportion of RNA in complex in the mix was determined by filter binding assay. The % of complex was plotted as a function of [RNA] equivalent of 40S, the linear part of the curve was calculated drawn, and extrapolated to 100% binding. The values are the mean of at least three independent experiments ± standard deviation. The two curves are statistically different (P < 10-4). (D) Analysis of the complexes formed by the full length HIV-1 Gag-IRES deleted of site 1 (Δ Site 1: A336–U851Δ363–485), of site 2 (Δ Site 2: A336–U851Δ632–756), or of both sites (Δ Site 1 Δ Site 2: A336–U851Δ363–485Δ632–756) and the full length HIV-1 Gag-IRES (A336–U851) with the 40S ribosomal subunit. 32P-labeled RNA transcripts were separated on 10–30% sucrose gradients. Peaks were identified by comparison with UV profiles obtained with purified 40S ribosomal subunits.