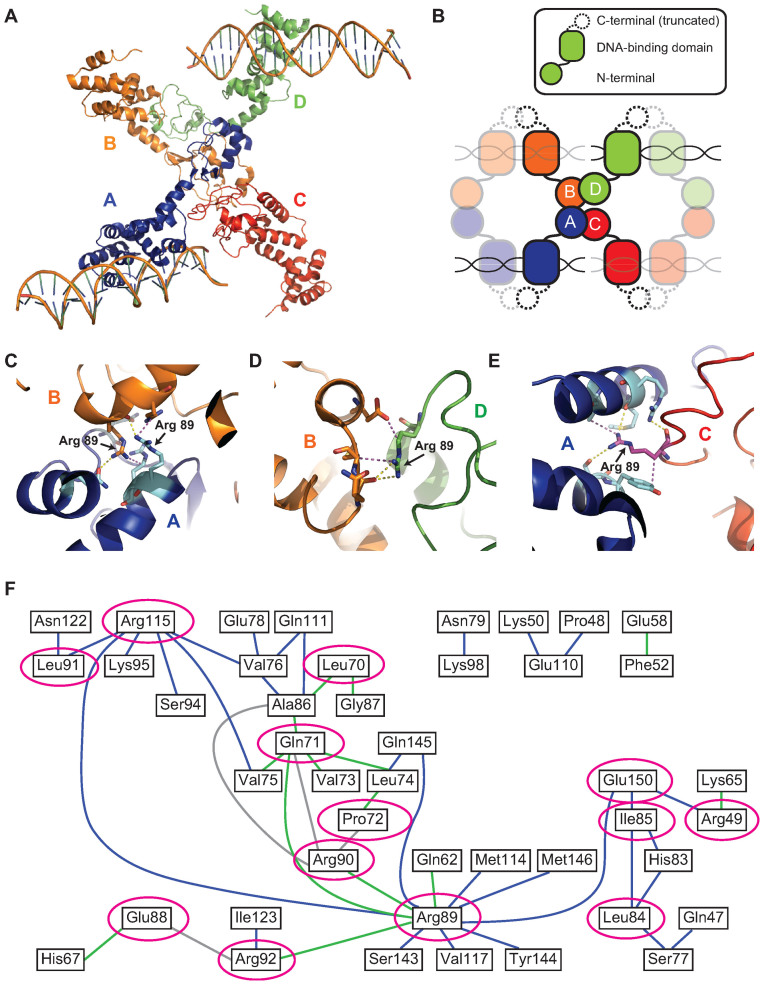
Figure 1.
Crystal structure of HpSpo0J reveals a network of cis and trans interactions for ParB spreading (see colors online). (A) Crystal structure of the C-terminally truncated HpSpo0J–parS complex (30) (PDB code: 4UMK). (B) Cartoon representation of the crystal structure (figure is not drawn to scale). Each chain (chain A in blue, chain B in orange, chain C in red and chain D in green) is a C-terminally truncated HpSpo0J monomer that is part of a dimer bound to half of a 24-bp parS DNA duplex. Only two DNA molecules were available from the original PDB file. Note that tetramerization of the HpSpo0J monomers is asymmetric due to chain C failing to interact with chain D in the crystal structure. (C) Multiple interactions in trans between chain A (blue) and chain B (orange) coordinated by the two R89 residues on each chain. (D) Multiple interactions in cis between chain B (orange) and chain D (green) coordinated by the single R89 residue on chain D. (E) Multiple interactions in cis between chain A (blue) and chain C (red) coordinated by the single R89 residue on chain C. Yellow dashed lines indicate hydrogen bonds, and magenta dashed lines indicate hydrophobic interactions. Figures were prepared in PyMOL. (F) A 2D network map generated from the crystal structure (see ‘Materials and Methods’ section) indicating cis (blue) and trans (green) interactions within the HpSpo0J–parS tetrameric complex. Interactions between residues within the same HpSpo0J monomer are shown in gray. Highly conserved residues that act as hubs for multiple interactions are circled in magenta. Residue number corresponds to that in HpSpo0J.