Abstract
In many next-generation sequencing (NGS) studies, multiple samples or data types are profiled for each individual. An important quality control (QC) step in these studies is to ensure that datasets from the same subject are properly paired. Given the heterogeneity of data types, file types and sequencing depths in a multi-dimensional study, a robust program that provides a standardized metric for genotype comparisons would be useful. Here, we describe NGSCheckMate, a user-friendly software package for verifying sample identities from FASTQ, BAM or VCF files. This tool uses a model-based method to compare allele read fractions at known single-nucleotide polymorphisms, considering depth-dependent behavior of similarity metrics for identical and unrelated samples. Our evaluation shows that NGSCheckMate is effective for a variety of data types, including exome sequencing, whole-genome sequencing, RNA-seq, ChIP-seq, targeted sequencing and single-cell whole-genome sequencing, with a minimal requirement for sequencing depth (>0.5X). An alignment-free module can be run directly on FASTQ files for a quick initial check. We recommend using this software as a QC step in NGS studies. Availability: https://github.com/parklab/NGSCheckMate
INTRODUCTION
Studies utilizing next-generation sequencing (NGS) technologies often involve comparison or integration of multiple datasets from a single individual. Different tissues or conditions from the same subject may be compared to identify tissue- or condition-specific mutations or transcriptional changes while controlling for genetic background, for example. In many cancer genome projects, tumor and matched normal genomes as well as their transcriptomes are sequenced for each patient to discover somatic mutations and their impact on gene expression. Other common situations include comparison of replicate experiments or merging of data from multiple lanes of a sequencer.
Correct labeling of the samples is essential for the integrity of downstream analysis. This quality control (QC) is particularly important in clinical applications, in which patient treatment may be informed by these data. Despite efforts to streamline sample-processing protocols, sample swapping can occur at various steps, especially when the tubes containing the samples are handled, for example during sample collection, DNA/RNA aliquot preparation, library construction or placement of the libraries on a sequencer. We have experienced the problem of inaccurately labeled samples in many projects, even with strict QC measures and high standards for data quality. Once a sample is found to have been mislabeled, data for that sample must be corrected or withdrawn. If the problem is detected in the late stages of analysis or even after publication, many analyses must be repeated, resulting in considerable loss of resources. Thus, one should perform sample-pairing QC of the sequencing data as early as possible in a study.
One approach to matching data to a particular individual is to examine short tandem repeats (STRs). The Cancer Genome Atlas (TCGA) uses a polymerase chain reaction-based assay to verify whether cancer and normal samples are derived from the same patient, targeting a handful of STRs from the CODIS database that are highly polymorphic among the human population (1). However, it is not feasible to genotype these CODIS STRs directly from sequencing data, especially whole-exome (WES) or RNA-seq data, because most of the STRs are located in non-coding regions or are longer than typical sequencing reads.
A more common approach is to use genotypes for single nucleotide polymorphisms (SNPs). Several studies proposed SNP-based sample checking for specific NGS data types such as RNA-seq (2) and WES (3). Another study has focused on checking across multiple lanes of sequencing data before combining them (4). The VCFtools software also provides options to verify kinship probabilities between samples based on variants in variant call format (VCF) files (5,6). However, all of the above methods are confined to a single data type and do not check for consistency across multiple data types. Some methods developed for microarray data examine the association between SNP genotype and either gene expression or DNA methylation to test sample pairing (7,8). Compared to microarray data, sequencing data enables improved genome-wide SNP genotyping in most data types, allowing direct comparison between genotypes rather than merely showing an association between genotypes and other features.
Here, we present NGSCheckMate, a stand-alone tool for sample-pairing QC, applicable to multiple data formats: the unaligned read format (FASTQ), the aligned read format (BAM) and the VCF. Our tool can be used to check pairings both between data of the same type and, importantly, between data of different types. For example, it can determine pairings between tumor and blood WES data and between RNA-seq and WES data from the same individual. The alignment-free module of NGSCheckMate can also be applied directly to raw sequencing data, avoiding the time-consuming alignment step. Our performance evaluation of NGSCheckMate using several data types with various sequencing depths shows that it is accurate and computationally efficient, making it a rapid and effective QC tool for a wide range of studies.
MATERIALS AND METHODS
Overview
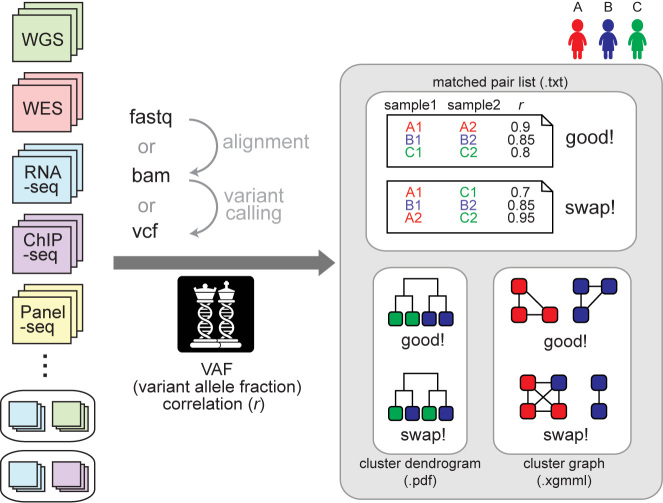
NGSCheckMate takes various types of NGS data in three formats (FASTQ, BAM or VCF) as input (Figure 1). It generates three types of output files (Figure 1): (i) a list of matched sample pairs with genotype correlation coefficients; (ii) a sample clustering dendrogram; and (iii) a graphical representation of sample clustering that can be entered into graphical visualization tools such as Cytoscape (9).
Figure 1.
A schematic overview. NGSCheckMate can handle various data types in any of the three formats (FASTQ, VCF or BAM). The tool calculates pairwise correlations of VAFs (variant allele fractions) from the input files and classifies each pair of files as either matched (from the same individual) or unmatched (not from the same individual). The output files are a text file listing the VAF correlation for each pair, a dendrogram image or an XGMML file with a graph structure that can be fed into graph visualization tools such as Cytoscape.
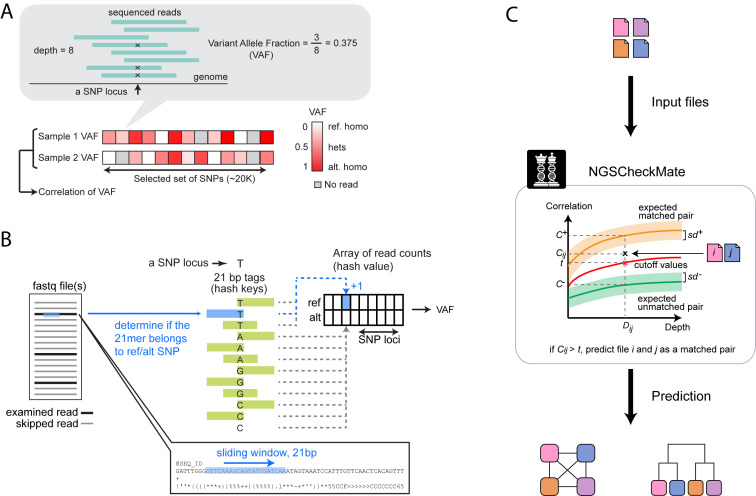
To determine whether two input files belong to the same individual, NGSCheckMate evaluates the correlation between the variant allele fractions (VAF) estimated from the two files at known SNPs (Figure 2A). The VAF of an SNP is defined as the ratio of the number of reads supporting a non-reference allele to the total number of reads spanning the SNP locus. For BAM input files, NGSCheckMate calculates VAFs using SAMtools mpileup (10) using the default options; for FASTQ input files, it scans the reads to search for k-mer sequences that span an SNP locus with a reference or alternative allele and calculates a VAF using the read counts containing the k-mer (Figure 2B; see ‘Materials and Methods’ section).
Figure 2.
Illustration of key steps. (A) A VAF is computed as the fraction of reads supporting a variant (non-reference) allele for a given SNP site. A VAF ranges between 0 and 1 at each genotype. The VAFs are computed across a panel of SNP sites for each file and the Pearson correlation between two VAF vectors is computed. (B) For the alignment-free module, a pre-built hash table stores 21-mer sequence tags that represent SNP sites and alleles. Each tag overlaps with the SNP site either at the center or at one of the two ends. For each SNP site, a total of 24 tag sequences (4 alleles × 3 overlapping SNP locations to the SNP site × 2 orientations (forward and reverse complementary)) are prepared and 6 and 18 of them represent a reference allele and alternative alleles, respectively. A hash is constructed with the 21-mer tags as keys, each pointing to an element of a 2-dimensional read count array, where the two dimensions are SNP loci and alleles. Given an input FASTQ file (single-end) or a pair of input FASTQ files (paired-end), randomly subsampled reads are examined by a 21-nt sliding window. If a 21-nt substring exists in the hash, we increase the corresponding read count value by one and move to the next read. In the end, VAFs are computed using the count values in the array. (C) A depth-dependent VAF correlation background model is constructed by down-sampling from high-coverage WGS data to 0.01–60X. Given input data files i and j, NGSCheckMate computes a VAF correlation coefficient Cij between the two files and compares it to the precomputed model at the observed depth Dij defined as the smaller of the mean depths for the two files. The VAF correlation cutoff for classification is the midpoint between the average correlation for matched pairs (C+) minus one standard deviation (sd+) and the average correlation for unmatched pairs (C−) plus one standard deviation (sd−) at a given depth.
A pair of data files is classified as matched or unmatched depending on whether their VAF correlation is closer to the pre-computed distribution of correlations for matched pairs from a training dataset or closer to the pre-computed distribution for unmatched pairs (Figure 2C; see ‘Materials and Methods’ section). Since the accuracy of SNP calls and thus the VAF correlations depend on sequencing depth, NGSCheckMate pre-computes and stores VAF correlations from datasets of diverse sequencing depths for comparison.
Selection of a reference SNP set
Among the SNP set (version 138) downloaded from dbSNP, we selected 21 067 exonic SNPs that are variable across individuals to construct a set of informative features for individual identification. Specifically, we calculated the VAF of every SNP in the dbSNP set using 40 germline WGS profiles from TCGA stomach cancer patients and selected SNPs whose median absolute deviation of the SNP VAF across samples is larger than zero. The resulting 21 067 exonic SNPs served as a reference set to measure VAF correlation between input files. For the alignment-free method, we required uniqueness of the k-mer sequence flanking the SNPs and used 11 696 SNPs (more details below). Our simulation showed that there was no difference in the distribution of VAF correlations between using the 21 067 and the reduced 11 696 SNP sets.
Alignment-free method
The alignment-free method is designed to obtain read counts for each SNP by scanning reads in FASTQ files to search for a k-mer sequence that spans the SNP locus either at the center or at one of the two ends (k = 21 for the current version). Both forward and reverse complementary sequences were included in the k-mer set. To ensure that each k-mer uniquely represents an SNP and its allele, we exclude SNPs for which the k-mers with the reference allele do not uniquely map to the reference genome or for which the k-mers with an alternative allele map to the reference genome. We used only perfect matches. Each k-mer (the hash key) derived from the remaining SNPs is stored in a hash table and points to the read count of the SNP and its allele type (the hash value). Every time we encounter a read with a k-mer in the hash, we increase the read count by one. Later, we use the read counts to calculate VAFs and perform the subsequent steps, as in the alignment-based method. To speed up the process, reads are randomly subsampled to a lower read depth that provides comparable accuracy.
Prediction
We estimated the distributions of VAF correlation coefficients for matched pairs (data files from the same individual) and unmatched pairs (data files from different individuals) using a training dataset of germline WGS data from 40 TCGA stomach cancer patients. To estimate the distributions at different sequencing depths, we subsampled reads from the original high coverage (>30X) data to 0.01–10X. Simulations varying factors such as the number of SNPs used, the read depth, and the read depth distribution showed that the estimated distributions were robust. We used the empirical distribution of VAF correlations as a reference model for prediction.
Given a pair of input files, we predicted that they are from the same person if their VAF correlation was closer to the VAF correlations for matched pairs than for unmatched pairs at similar sequencing depths. Practically, we divided sequencing depths into multiple intervals and pre-calculated an interval-specific VAF correlation cutoff, roughly the midpoint between the two distributions of VAF correlations for matched and unmatched pairs, as illustrated in Figure 2C. The cutoffs are 0.38, 0.41, 0.46, 0.55 and 0.61 for unrelated data with sequencing depth of <1, [1, 2), [2, 5), [5, 10) and ≥10, respectively. For related data including family data, the cutoffs were 0.50, 0.54, 0.59 0.69 and 0.76, respectively. For WGS, WES and RNA-seq data, the mean depth across all of the 20K or 12K exonic SNPs was used as a reference depth to retrieve the corresponding VAF correlation cutoff. For Panel-seq and ChIP-seq data where many SNPs do not have mapped reads, the mean depth across a subset of the SNPs with at least one mapped read (the mean of non-zero depths) was used as a reference depth to determine the VAF correlation cutoff. When the reference sequencing depths from input files span different intervals, we used the lower sequencing depth to find the corresponding interval and the VAF correlation cutoff. For example, if an input file A has an average sequencing depth of 3X, and another input file B has an average sequencing depth of 7X, then we predict files A and B to be from the same person when their VAF correlation coefficient is larger than 0.46, the cutoff value for the depth interval [2,5). For datasets with samples of related individuals (e.g. parent-child and siblings), more stringent VAF correlation cutoff values were used to distinguish matched from unmatched pairs.
Preparing WGS data of various sequencing depths
To construct a depth-dependent prediction model, we generated WGS data of various sequencing depths by splitting reads from different sequencing lanes or by subsampling reads from the original high-depth data. We used the splitBam function of BamUtil (http://genome.sph.umich.edu/wiki/BamUtil) to separates reads per sequencing lane annotated by RG tags in BAM files. From 18 pairs of TCGA stomach cancer WGS data (35–74X), we obtained 147 BAM files of sequencing depth 1–25X separated by sequencing lanes. For down-sampling, we used samtools view –s sampling_ratio, where the sampling_ratio was used to achieve the desired read depth. We generated WGS data of depth (X) 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 2, 5, 10 and 30, and WES data of depth (X) 0.5, 1, 2, 5, 10 and 30.
Simulation
Simulation was performed to test the effect of various factors on VAF correlations in identical, related and unrelated individuals. First, an SNP was simulated by randomly drawing two alleles from a distribution of allele frequencies in the human population. Each individual was then represented by n independently generated SNPs. We tested n = 10 000 and n = 20 000, to address alignment-free and post-alignment SNP sets, respectively. We assumed a uniform distribution of variant allele frequencies, as reported in the HapMap study (11). We also tested an alternative distribution (Supplementary Figure S1C, β distribution with a = 1, b = 2). The uniform distribution produced a set of SNPs where a third were heterozygotes, a third were non-reference homozygotes and the other a third were reference homozygotes, and the ratio of heterozygotes to non-reference homozygotes was 1.0. In contrast, the β distribution resulted in a set of SNPs where ∼35% were heterozygotes, ∼16% were non-reference homozygotes and the ratio of heterozygotes to non-reference homozygotes was ∼2.2. Both distributions showed consistency with the previous reports that the proportion of heterozygotes was ∼35% (12) and that the ratio of heterozygotes to non-reference homozygotes ranged 1–2.2 (13).
Two unrelated individuals were simulated by independently drawing two individuals as described above. To simulate a parent-child pair or siblings, we randomly drew an allele for each SNP from each of two unrelated individuals (parents) to represent a child genotype.
To simulate VAFs, we considered the read depth distribution and the distribution of fraction of reads supporting an alternative allele. Since the non-uniformity of depth in the sequencing data can adversely affect the performance by reducing the number of informative SNPs, we simulated the depth distribution to be similar to that of real WGS data. To simulate the total depth for each SNP site, we first trained a series of negative binomial (NB) models from the depths across the 21 067 SNP sites using the WGS data from TCGA stomach cancer, colorectal cancer and lymphoma samples, including the original and down-sampled ones. The estimated inverse of the shape parameter (1/r) ranged between 0 and 5 when the average depth was larger than 0.05X. The larger the variance of the depth, the smaller the inverse shape parameter (equivalent to Poisson variance when the shape parameter reaches infinity). For each set of simulations, the depth distribution was chosen to be either a Poisson or an NB with 1/r = 5. For each SNP site, we independently drew a depth from the chosen depth distribution. Then, to simulate fraction of reads supporting alternative alleles, we used by default a binomial model with p = 0.5 for each heterozygous site. For comparison, we also used a uniform distribution that reflects higher allelic imbalance, as often observed in single-cell data. The mean and 5 and 95% quantiles of VAF correlations were derived from 1000 replicate simulations for each category. We used the fitdistrplus (14) R package to train a NB model. Some of the factors used in simulation are described in Supplementary Figure S1.
Copy number variation (CNV) analysis
To further investigate whether samples with different phenotypes or disease status (e.g. cancer versus normal) are from the same person, we utilized germline copy number variations (CNVs). Briefly, we identified CNVs using the read-depth-based BIC-seq2 algorithm, a revised version of BIC-seq (15) for analyzing WGS data without matched controls (16). We selected CNVs that overlap with known germline CNVs reported in the Database of Genomic Variants Gold Standard CNV set (17), with at least 50% reciprocal overlap. This filtering reduces false discovery rate in our CNV calls. To reduce false omission rate (an unobserved site being a real CNV) and thus improve our comparison accuracy, we used only those CNVs with population frequency lower than 50% (using the ‘nr_frequency’ column).
Datasets tested
We evaluated our method using data generated from TCGA, Samsung Genome Institute and previous studies (18–21) (Supplementary Table S1). The TCGA data we downloaded from cgHub (http://cghub.ucsc.edu/) included 106 WGS pairs (cancer and matched normal tissue or blood) from stomach and colorectal cancer patients, 984 WES pairs from 10 cancer types and 108 RNA-seq profiles from stomach and colon cancer samples. All TCGA data are available for download by users with data access approval at the NCI Genomic Data Commons Portal (https://gdc-portal.nci.nih.gov/); its accession number at the Database of Genotypes and Phenotypes (dbGaP) is phs000178.v8.p7. More information about TCGA can be found at http://cancergenome.nih.gov. The data generated from the Samsung Genome Institute included 14 pairs of lymphoma WGS, 68 pairs of breast cancer WES, 53 profiles of RNA-seq and 85 profiles of panel-sequencing from 34 patients of 5 cancer types (brain, kidney, colon, breast and lung cancers). The panel sequencing captured ∼80-400 cancer-associated genes at sequencing depth of >800X from cancer (primary and metastasis tumors, or biopsies of multiple regions of the tumor) and normal tissue samples. We also tested our method using two single-cell WGS datasets: (i) 36 high-coverage (average depth ∼42X) single-neuronal WGS profiles from three post-mortem brains of neurotypical individuals (16, 10 and 10 cells for each individual) (20), and (ii) 89 single-cell WGS data (0.01–0.3X) from two glioblastoma patients (39 profile from BT325 and 50 from BT340) (18). One profile (SRR1779165) was removed because its coverage was too low (0.0001X) to examine SNPs. We also analyzed ChIP-seq profiles for CTCF, H3K4me1, H3K4me3, H3K27ac, H3K27me3, H3K36me3 and SA1 from eigth healthy individuals (GM18505, GM18486, GM19099, GM2255, GM18951, GM19193, GM18526, GM19240) downloaded from SRA (http://www.ncbi.nlm.nih.gov/sra; SRP030041) (19). Each individual had input DNA and two or three replicate ChIP-seq profiles for each histone mark or transcription factor. All except one individual had RNA-seq profiles.
In addition to all the Illumina data above, we tested our method on Ion Torrent data. Specifically, the exome data files from the two individuals (NA12878 and NA24631) were downloaded from the Genome In a Bottle Consortium consortium site (ftp://ftp-trace.ncbi.nih.gov/giab/ftp/data/) (21). The BAM files were subsampled to simulate lower-coverage data using Samtools v1.2. Variants were then called on these sub-sampled BAM files with Torrent Variant Caller 5.0-2 at default settings, except that the minimum coverage for SNP calling was set to 1.
RESULTS AND DISCUSSION
Robust separation of VAF correlations between matched and unmatched pairs
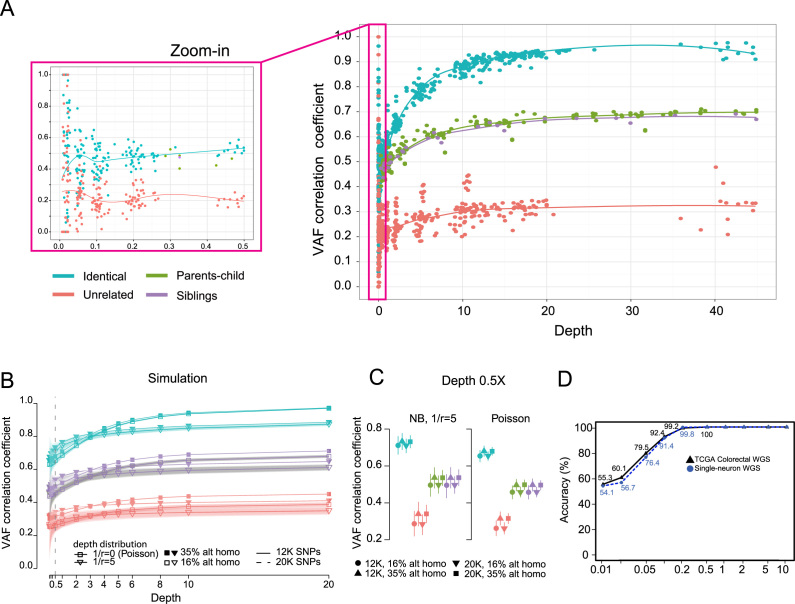
When the two pre-computed distributions of VAF correlation coefficients for matched versus unmatched pairs are far apart, our prediction becomes more reliable. To empirically estimate the distribution of VAF correlations, we used the germline WGS data from 40 TCGA stomach cancer patients (22), with subsampling at multiple depths starting at 0.01X. The VAF correlations of matched pairs were clearly separable from those of unmatched pairs for depths as low as 0.5X (Figure 3A). To determine whether VAF correlations of related individuals (parent-child or siblings) differ from those of the same individuals, we examined 36 WES profiles obtained from ten families. The VAF correlations of the related pairs (parent-child and sibling pairs) were distinct from those of the matched pairs for depths as low as 3X (Figure 3A).
Figure 3.
Classification based on VAF correlations. (A) Depth-dependent VAF correlations derived from WGS and WES datasets for different types of sample pairs: identical, related (parent-child or siblings) and unrelated. The LOESS regression lines are shown. Family pairs were tested only for depths >0.5X. (B) VAF correlations based on simulation, with different shape parameters (r) of a negative binomial model for the read depth distribution, numbers of SNPs used (12K versus 20K), and percentages of alternative homozygous SNPs (see ‘Materials and Methods’ section). A binomial distribution was assumed for the distribution of alternative allele reads (see ‘Materials and Methods’ section). The top and bottom 5% of simulated VAF correlations are plotted as shaded areas around each line. VAF correlations of parent-child pairs (green) are hidden in the figure because they overlap those of sibling pairs (purple). VAF correlations using 12K (solid lines) and 20K SNPs (dotted lines) are also indistinguishable (nearly superimposed in the figure). (C) The distributions of simulated VAF correlations in (B) at the depth of 0.5X are shown in vertical lines representing the top and bottom 5% VAF correlation values. (D) Accuracy at various sequencing depths for WGS data. The original and down-sampled datasets of 66 TCGA colorectal pairs and 36 single-neurons from three post-mortem brains were tested.
To understand the factors that may affect the VAF correlation distribution, we performed simulations to analyze the effects of several parameters such as the number of SNPs used, read depth and the fraction of homozygous SNPs (see ‘Materials and Methods’ section). The distributions of simulated VAF correlations for matched and unrelated sample pairs showed a clear separation at a depth as low as 0.5X (Figure 3B and C). The matched and family pairs could also be easily distinguished even at very low depth.
Performance of NGSCheckMate
We evaluated the performance of the method for WGS, WES, targeted sequencing for selected loci (‘panel-seq’), RNA-seq and ChIP-seq. The testing data included 160 WGS, ∼1000 WES, ∼170 RNA-seq, 85 panel-seq, ∼130 ChIP-seq and ∼130 single-cell WGS data files (Table 1; see ‘dataset’ in ‘Materials and Methods’ section for details). The majority of WGS, WES and RNA-seq files were obtained from the TCGA project; the rest of these files (including panel-seq) were obtained from various projects at the Samsung Genome Institute. The ChIP-seq files for input DNA, histone modifications and transcription factors from eight individuals were obtained from a published study (19). Single-cell WGS files from post-mortem human brains of three neurologically normal individuals and two glioblastoma patients were obtained from two previous studies (18,20).
Table 1. NGSCheckMate performance.
| Data type | Dataset (pair type) | Sequencing depth1 | Individual | Sample | Test pairs #matched, #unmatched | Accuracy (%)2 |
|---|---|---|---|---|---|---|
| WGS (BAM) | TCGA colorectal (cancer versus normal) | >30X, down-sampling (0.5–30X) | 66 | 132 | 66, 66 | 100 |
| down-sampling (0.01–0.2X) | 55.3–99.2 | |||||
| WGS (BAM, FASTQ) | non-TCGA lymphoma (cancer versus normal) | 30–60X, down-sampling (0.5–30X) | 14 | 28 | 14, 28 | 100, 100 |
| WES (BAM) | TCGA 9 cancer types (cancer versus normal) | ∼100X | 421 | 842 | 421, 421 | 100 |
| TCGA kidney (cancer versus normal) | ∼100X, down-sampling (0.5–30X) | 50 | 100 | 50, 50 | 100 | |
| WES (FASTQ) | non-TCGA breast (cancer versus normal) | ∼60X, down-sampling (0.5–10X) | 68 | 136 | 68, 68 | 100 |
| RNA-seq (BAM) | TCGA colorectal (cancer versus normal) | ∼65X, down-sampling (0.5–10X) | 19 | 38 | 19, 19 | 100 |
| Single-cell WGS (BAM) | single-neuron | ∼42X, down-sampling (0.5–10X) | 3 | 36 | 210, 210 | 100 |
| glioblastoma (cancer–cancer) | 0.01–0.3X | 2 | 89 | 45, 45 | 87.8 | |
| Chip-seq (BAM, FASTQ) | within marks | 5.4 (2.2–19.0) | 8 | 119 | 72, 72 | 97.6, 97.7 |
| input versus mark | input DNA 2.3 (2.1–2.9) | 8 | 127 | 133, 133 | 98.5, 99.8 | |
| Panel-seq (BAM, FASTQ) | cancer versus normal, multiple regions, primary versus metastasis | 40 (20–119) | 5, 18, 11 | 12, 48, 25 | 92, 87 | 98.3, 99.4 |
| RNA-seq versus WES (BAM) | TCGA stomach (cancer or normal DNA versus cancer RNA) | RNA-seq (∼70X) WES (∼100X) | 65 | 201 | 132,132 | 100 |
| RNA-seq versus WES (FASTQ) | non-TCGA breast cancer (cancer or normal DNA versus cancer RNA) | RNA-seq (∼25X) WES (∼60X), down-sampling for both datasets (0.5–10X) | 53 | 159 | 106, 106 | 100 |
| RNA- versus ChIP-seq (BAM, FASTQ) | RNA-seq versus ChIP-seq (all marks) | RNA-seq (∼5X) ChIP-seq (described above) | 7 | 119 | 231, 231 | 99, 98.9 |
1For WGS, WES and RNA-seq, the average mean depth is shown. For Panel-seq and ChIP-seq, the average of the mean non-zero depths across the SNPs with at least one mapped read is shown with its range in parentheses.
2Accuracy estimates for the alignment-based and the alignment-free method are separated by a comma.
Within-platform comparisons
For each test pair, we selected files from the same individual (‘matched’) or from different individuals (‘unmatched’). We applied NGSCheckMate to all of the test pairs to predict whether each pair was from the same individual or not. We compared the predictions to our known labels and calculated the accuracy as the percentage of correctly predicted pairs. The number of possible unmatched pairs is larger than the number of matched pairs, so we generated up to ten sets of randomly selected unmatched pairs of similar size to matched pairs and reported the average accuracy across the multiple unmatched pair sets.
For WGS, WES and RNA-seq data, both alignment-free (FASTQ input files) and alignment-based (BAM or VCF input files) methods correctly predicted all pairs when the sequencing depth was at least 0.5X (Figure 3D and Table 1). Genotyping individual SNPs at a shallow sequencing depth is unreliable, but NGSCheckMate achieved a high degree of accuracy in matching samples by combining signals (variant allelic frequencies) from a large number of SNPs. Not surprisingly, the performance diminished at a very shallow depth (<0.5X), as noise starts to dominate the correlation (Figure 3D).
We tested our method on two single-cell WGS datasets (18,20) to discriminate single cells from different individuals. One dataset consisted of high-coverage (∼42X) 36 WGS profiles of single neuronal genomes from three postmortem brains. Our method on this whole genome amplified dataset showed comparable performance to the typical unamplified WGS datasets, achieving 100% accuracy at a sequencing depth >0.5X (Figure 3D and Table 1). The other dataset consisted of 89 WGS profiles of single cancer cells from two glioblastoma patients (39 and 50 cells from each patient), sequenced at a very low depth (0.01–0.3X) to characterize CNV at the single cell level. Our evaluation showed 87.8% accuracy in grouping the cells, with all misclassification errors due to a few cells with especially shallow sequencing depth (<0.15X).
In predicting whether two ChIP-seq profiles (histone modification or transcription factor) were from the same individual, the accuracy was 97.6% for the alignment-based method and 97.7% for the alignment-free method (Table 1 for across mark and Supplementary Table S2 for per mark performance). We also tested whether input DNA and ChIP-seq profiles were from the same individual, and obtained 98.5 and 99.8% accuracy for the alignment-based and the alignment-free method, respectively. Although only a tiny fraction of the genome is covered in most ChIP-seq profiles, they still covered a sufficient number of SNPs to allow for accurate prediction.
Finally, the panel-seq platform covered a subset of cancer-associated genes (80–400 genes) with very high coverage (>800X). The 85 samples we tested included cancer/normal tissue pairs, primary tumor/metastasis pairs and multiple regions of the same tumors. NGSCheckMate correctly predicted 98.3 and 99.4% of the tested matched and unmatched pairs, using BAM or FASTQ files, respectively (Table 1).
Cross-platform comparisons
Many projects generate multiple types of data for each individual. For example, TCGA has generated WGS and/or WES as well as RNA-seq data and others from the same patient; Roadmap Epigenomics has generated RNA-seq and ChIP-seq profiles for each tissue or cell line. To show that NGSCheckMate identifies correct pairs across data types, we tested grouping RNA-seq and WES datasets from the same individuals. Our results showed 100% accuracy even when both datasets were down-sampled to 0.5X (Table 1). We also tested if NGSCheckMate could pair RNA-seq and ChIP-seq profiles from the same individual. This test achieved 99 and 98.9% accuracy for the alignment-based and the alignment-free method, respectively (Table 1 and Supplementary Table S2).
Effect of allelic imbalance
We also evaluated the impact of allelic imbalance in transcriptomic and epigenetic data due to monoallelic expression on the performance of our method. We collected 4227 genes with monoallelic expression from a published study (23) and removed the 4241 SNPs contained within those genes from the 21 067 SNPs. In all cases, the performance with and without the 4241 SNPs were very similar: no difference for RNA-seq versus RNA-seq at all sequencing depths (100% accuracy for 0.5X to original depth); ∼0.7% difference for pairs from ChIP-seq marks; and ∼0.3% difference for RNA-seq versus ChIP-seq. Overall, this analysis shows that allelic imbalance has little impact on the performance of NGSCheckMate.
Non-Illumina platforms
We tested whether our pre-built model derived from the Illumina sequencing platform would be generalizable to non-Illumina sequencing data. Using the exome sequencing data generated on the Ion Torrent platform for two individuals, we generated multiple down-sampled BAM files from each original BAM file and confirmed that our method was able to accurately distinguish matched pairs from unmatched pairs at different depths (0.5, 1, 2, 5 and 10X down-sampled datasets). Although it would require tests on more samples and other platforms to claim generalizability, this preliminary test shows that the combined signals from numerous SNPs may be robust enough to overcome heterogeneous error profiles across platforms.
Comparison with other genotype-based methods
Some NGS studies generate genotype calls as part of their analytical pipelines, and the genotypes instead of VAFs could serve as features for sample-pairing QC. Thus, we tested how our method using VAF correlation compares to two approaches based on the genotype calls at the same ∼20K known SNPs: (i) the correlation between variant genotypes and (ii) the fraction of concordant genotypes between two samples. Typical genotype-calling methods such as GATK and samtools/vcftools report only non-reference variants (heterozygous or alternative homozygous variants). In our comparison, we tested methods with three genotypes where non-genotyped SNPs are considered as reference homozygous variants (Supplementary Figure S4) as well as with two non-reference genotypes only (Supplementary Figure S5). Overall, all three measures show good separation between matched and unmatched pairs. However, the VAF correlation shows greater separation between the correlation curves than for other measures when the depth is very low (< 0.5X and < 2.5X when using three and two genotypes, respectively) (Supplementary Figures S5 and 6). Furthermore, the VAF correlation thresholds obtained from training WGS data (green lines) were equally applicable to other data types, whereas the correlation thresholds for the three genotype-based metrics were highly variable among different data types (Supplementary Figure S5). This might be because different types of non-WGS data types have variable numbers of SNPs that have no or few mapped reads. Our method based on VAF correlation is more robust to variations in parameters in genotype calling and is generalizable to other data types.
Use cases
Here, we illustrate the utility of NGSCheckMate by describing several studies in which we identified potentially mislabeled samples.
Liver cancer WGS project
In this project, we generated WGS (>30X) data for 21 liver cancer patients, with three samples per patient (cancer, adjacent normal tissue and blood). While analyzing somatic single nucleotide variants (SNVs), we noticed that different patients shared an unusually large number of somatic SNVs. This was unexpected, as the majority of somatic SNVs should be specific to individual tumors. A further investigation found that a systematic sample mislabeling had occurred at the sequencing center when the tubes were labeled. NGSCheckMate identified all of the mislabeled samples. After we resequenced the problematic samples, NGSCheckMate produced a correct clustering, with three samples per patient.
Metastatic colon cancer project
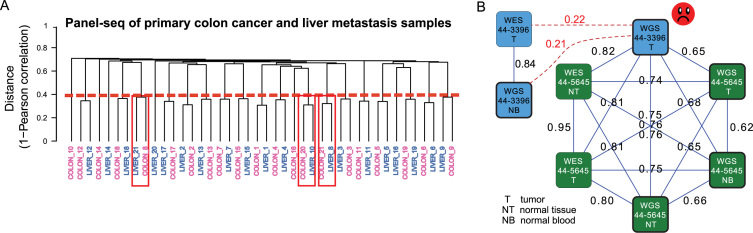
We generated targeted sequencing data for 21 primary colon cancers and their liver metastasis counterparts to understand genomic alterations that take place during metastasis. NGSCheckMate identified several unexpected sample clusters, as shown in the dendrogram in Figure 4A. For example, patients 8 and 21 had either their primary samples or their metastasis samples swapped with each other. There were additional samples (e.g. patients 10 and 20) that did not show proper sample matching. We examined these problematic samples in detail and excluded them from further analysis when the correct labeling could not be resolved.
Figure 4.
Examples of sample mislabelings. (A) A dendrogram output for panel-seq data of primary colon cancer and liver metastasis samples, based on 1 - VAF correlation as the distance measure. The numbers in the sample names are the patient IDs. The red line indicates the average of VAF correlation cutoffs used for predicting matching status of each pair; the VAF correlation cutoff used for every pair depends on the smaller of the depths for the two input profiles. Since this is panel-sequencing data, the mean of non-zero depths was used as the reference depth to retrieve a corresponding VAF correlation cutoff from the pre-built model (see Methods for details). Samples predicted to be mislabeled with high confidence were marked by red boxes. (B) Part of a graph output for the TCGA lung cancer WGS (low-coverage) and WES datasets. A node represents an input file, colored by individual. A solid edge represents a matched pair predicted by NGSCheckMate. The corresponding VAF correlation is written next to each edge. The graph indicates a mislabeling of 44–3396 T (tumor) WGS file. VAF Correlations between the 44–3396 T file and each of the other two 44–3396 files (tumor WES and blood WGS) did not pass a VAF correlation cutoff for a matched pair (red dotted lines and texts).
Low-coverage WGS data in TCGA (stomach)
TCGA provided low-coverage WGS data for a subset of patients for structural variation analysis. We analyzed the sample pairing for those patients with stomach cancer. Among the 242 BAM files in the stomach cancer dataset, NGSCheckMate identified one potentially mislabeled BAM file (a blood sample from patient TCGA-CG-4301) that did not match the two other BAM files labeled as coming from the same patient. In fact, TCGA had previously identified ∼10% of their stomach cancer samples as mislabeled at the TCGA aliquot distribution site by comparing their genotypes called by GATK, after an investigation was triggered by the detection of several identical fusions in different cancer patient samples. All mislabeled files had been redacted from the TCGA data repository (cgHub) except for the file that NGSCheckMate identified as mislabeled.
Low-coverage WGS data in TCGA (lung and other cancer types)
We also examined the TCGA lung adenocarcinoma WGS dataset. Among the 252 BAM files at cgHub, NGSCheckMate detected several potentially problematic files. To investigate this problem further, we ran NGSCheckMate on a larger dataset that included both WGS and WES data from the same individuals. In Figure 4B, we illustrate the case in which the tumor WGS file from individual 44–3396 matched with three WGS and two WES files, all from individual 44–5645, instead of matching to the tumor WES or blood WGS files from individual 44–3396. This result strongly suggested that the tumor WGS file labeled ‘44–3396’ is likely to be from individual 44–5645. Additional analysis based on germline CNV also supported this hypothesis (Supplementary Figure S2).
We also performed a comprehensive screening for sampling swapping in additional 2316 TCGA low-coverage WGS BAM files across 14 cancer types and identified potential sample mislabelings in 5 additional cancer types (Supplementary Table S3). We are currently investigating these cases with the data generation group and are seeking a way to annotate data quality on the new TCGA data portal.
Speed and memory requirements
Since a large number of files may need to be checked, speed and low memory usage for the algorithm are important. Read alignment to generate BAM files may take hours or even days for large FASTQ files. Once the BAM files are available, the next time-consuming step in the alignment-based module is SAMtools mpileup. For a 30X WGS BAM file, the mpileup process takes about an hour to generate a VCF file for 21 067 SNPs using Intel(R) Xeon(R) CPU L5640 2.27GHz with 1 core and 4 GB memory. However, this can be sped up considerably by breaking up the BAM file into smaller pieces and running mpileup on them in parallel. For a WES BAM file (or other BAM files of similar file size), the mpileup process takes less than one minute using the same SNP set. Once VCF files are generated, pairwise correlation coefficients can easily be generated; for example, 80 WGS VCF files can be processed in ∼3 minutes using less than 200 MBs of memory.
To avoid the alignment step all together, we also developed an alignment-free module (see ‘Materials and Methods’ section). Here, the slow step is reading the input FASTQ files, but we provide a parallelization option using multiple cores where each core reads a different partition of a FASTQ input file. To further reduce run time, we offer an option to randomly subsample reads, given our performance evaluation that showed similar performance at lower depths down to 2X. Our tests show that the run time is near-linear with respect to both the number of cores and the total number of subsampled reads (Supplementary Figure S3). Our benchmarking test for a pair of RNA-seq FASTQ files (17 million 101 bp reads) took less than one minute with a read subsampling to ∼2X and a single core. Our test for a pair of WGS FASTQ files (∼300 million 101 bp reads) took 11 minutes with a read subsampling to ∼2X and 8 cores. The memory used was less than 40MB in both cases.
CONCLUSION
We have developed NGSCheckMate to identify datasets that belong to the same individual and have demonstrated its effectiveness in identifying potentially mislabeled datasets within and across diverse data types. It achieves excellent performance even for datasets with sequencing depths as low as 0.5X and with limited genomic coverage such as targeted sequencing and ChIP-seq data. The alignment-free version is also available for mislabeling detection early in an analysis pipeline. We recommend NGSCheckMate as an important part of the analysis pipeline in studies that involve multiple samples/files per subject.
Supplementary Material
ACKNOWLEDGEMENTS
The results published here are in part based upon data generated by TCGA managed by the NCI and NHGRI. Information about TCGA can be found at http://cancergenome.nih.gov. We thank Semin Lee for helpful discussions and sharing TCGA germline CNV profiles, Fritz Lekschas for designing the NGSCheckMate logo and members of the Park laboratory including Maxwell Sherman, Euncheon Lim, Alison Barton, Su Wang and Peter Kerpedjiev for providing feedback on the manuscript and/or testing the NGSCheckMate program. We thank Nayoung K. D. Kim, Hyun-Tae Shin and Jae Won Yun at Samsung Medical Center for providing information on their clinical samples. We also thank Korea Telecom and Supercomputing Center of the Korea Institute of Science and Technology Information (Hyojin Kang and Junehawk Lee) for providing computing resources and technical support.
Authors’ contributions: E.A.L. conceived the initial idea for the project and the software. Se.L., So.L. and E.A.L. designed the study, developed the method/software and analyzed the data. S.O. analyzed the TCGA low-coverage WGS data. All authors participated in the writing of the manuscript. E.A.L. and P.J.P. provided supervision. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Harvard Medical School Eleanor and Miles Shore Fellowship [to E.A.L.]; Randolph Hearst Fund [to E.A.L.]; National Institute on Aging [K01AG051791 to E.A.L.]; National Institute of Mental Health [1P50MH106933, 1U01MH106883 to P.J.P.]; National Eye Institute [R01EY024230 to P.J.P.]; Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea [HI13C2096 to W-Y.P.]. Funding for open access charge: NIH [1U01MH106883].
Conflict of interest statement. None declared.
REFERENCES
- 1. Katsanis S.H., Wagner J.K.. Characterization of the standard and recommended CODIS markers. J. Forensic Sci. 2013; 58(Suppl. 1):S169–S172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang J., Chen J., Lathrop M., Liang L.. A tool for RNA sequencing sample identity check. Bioinformatics. 2013; 29:1463–1464. [DOI] [PubMed] [Google Scholar]
- 3. Pengelly R.J., Gibson J., Andreoletti G., Collins A., Mattocks C.J., Ennis S.. A SNP profiling panel for sample tracking in whole-exome sequencing studies. Genome Med. 2013; 5:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goldfeder R.L., Parker S.C., Ajay S.S., Ozel Abaan H., Margulies E.H.. A bioinformatics approach for determining sample identity from different lanes of high-throughput sequencing data. PLoS One. 2011; 6:e23683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. et al. . The variant call format and VCFtools. Bioinformatics. 2011; 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maurano M.T., Haugen E., Sandstrom R., Vierstra J., Shafer A., Kaul R., Stamatoyannopoulos J.A.. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet. 2015; 47:1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoo S., Huang T., Campbell J.D., Lee E., Tu Z., Geraci M.W., Powell C.A., Schadt E.E., Spira A., Zhu J.. MODMatcher: multi-omics data matcher for integrative genomic analysis. PLoS Comput. Biol. 2014; 10:e1003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Broman K.W., Keller M.P., Broman A.T., Kendziorski C., Yandell B.S., Sen S., Attie A.D.. Identification and correction of sample mix-ups in expression genetic data: a case study. G3 (Bethesda). 2015; 5:2177–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cline M.S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B. et al. . Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007; 2:2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. International HapMap, C. A haplotype map of the human genome. Nature. 2005; 437:1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderson C.A., Pettersson F.H., Clarke G.M., Cardon L.R., Morris A.P., Zondervan K.T.. Data quality control in genetic case-control association studies. Nat. Protoc. 2010; 5:1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kidd J.M., Gravel S., Byrnes J., Moreno-Estrada A., Musharoff S., Bryc K., Degenhardt J.D., Brisbin A., Sheth V., Chen R. et al. . Population genetic inference from personal genome data: impact of ancestry and admixture on human genomic variation. Am. J. Hum. Genet. 2012; 91:660–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dutang M.L.D.-M.a.C. fitdistrplus: an R package for fitting distribution. J. Stat. Softw. 2015; 64:1–34. [Google Scholar]
- 15. Xi R., Hadjipanayis A.G., Luquette L.J., Kim T.M., Lee E., Zhang J., Johnson M.D., Muzny D.M., Wheeler D.A., Gibbs R.A. et al. . Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:E1128–E1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xi R., Lee S., Xia Y., Kim T.M., Park P.J.. Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants. Nucleic Acids Res. 2016; 44:6274–6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. MacDonald J.R., Ziman R., Yuen R.K., Feuk L., Scherer S.W.. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014; 42:D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Francis J.M., Zhang C.Z., Maire C.L., Jung J., Manzo V.E., Adalsteinsson V.A., Homer H., Haidar S., Blumenstiel B., Pedamallu C.S. et al. . EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014; 4:956–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kasowski M., Kyriazopoulou-Panagiotopoulou S., Grubert F., Zaugg J.B., Kundaje A., Liu Y., Boyle A.P., Zhang Q.C., Zakharia F., Spacek D.V. et al. . Extensive variation in chromatin states across humans. Science. 2013; 342:750–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lodato M.A., Woodworth M.B., Lee S., Evrony G.D., Mehta B.K., Karger A., Chittenden T.W., D’Gama A.M., Cai X., Luquette L.J. et al. . Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015; 350:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zook J.M., Chapman B., Wang J., Mittelman D., Hofmann O., Hide W., Salit M.. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol. 2014; 32:246–251. [DOI] [PubMed] [Google Scholar]
- 22. Bass A., Thorsson V., Shmulevich I., Reynolds SM., Miller M., Bernard B., Hinoue T., Laird PW., Curtis C., Shen H. et al. . Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014; 513:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Savova V., Chun S., Sohail M., McCole R.B., Witwicki R., Gai L., Lenz T.L., Wu C.T., Sunyaev S.R., Gimelbrant A.A.. Genes with monoallelic expression contribute disproportionately to genetic diversity in humans. Nat. Genet. 2016; 48:231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.