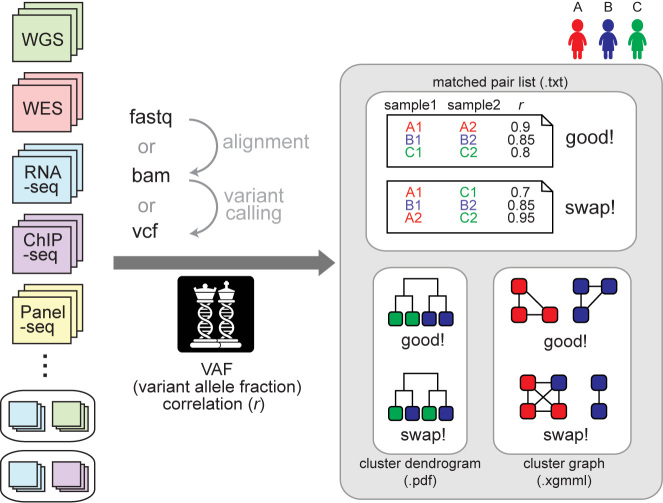
Figure 1.
A schematic overview. NGSCheckMate can handle various data types in any of the three formats (FASTQ, VCF or BAM). The tool calculates pairwise correlations of VAFs (variant allele fractions) from the input files and classifies each pair of files as either matched (from the same individual) or unmatched (not from the same individual). The output files are a text file listing the VAF correlation for each pair, a dendrogram image or an XGMML file with a graph structure that can be fed into graph visualization tools such as Cytoscape.