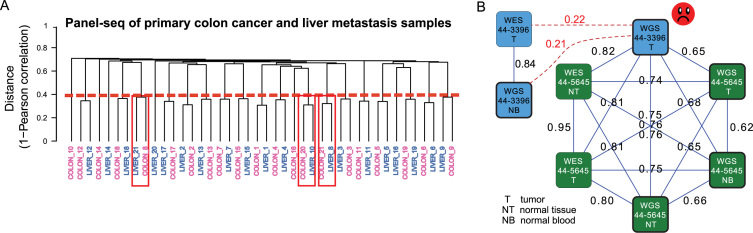
Figure 4.
Examples of sample mislabelings. (A) A dendrogram output for panel-seq data of primary colon cancer and liver metastasis samples, based on 1 - VAF correlation as the distance measure. The numbers in the sample names are the patient IDs. The red line indicates the average of VAF correlation cutoffs used for predicting matching status of each pair; the VAF correlation cutoff used for every pair depends on the smaller of the depths for the two input profiles. Since this is panel-sequencing data, the mean of non-zero depths was used as the reference depth to retrieve a corresponding VAF correlation cutoff from the pre-built model (see Methods for details). Samples predicted to be mislabeled with high confidence were marked by red boxes. (B) Part of a graph output for the TCGA lung cancer WGS (low-coverage) and WES datasets. A node represents an input file, colored by individual. A solid edge represents a matched pair predicted by NGSCheckMate. The corresponding VAF correlation is written next to each edge. The graph indicates a mislabeling of 44–3396 T (tumor) WGS file. VAF Correlations between the 44–3396 T file and each of the other two 44–3396 files (tumor WES and blood WGS) did not pass a VAF correlation cutoff for a matched pair (red dotted lines and texts).