Figure 3.
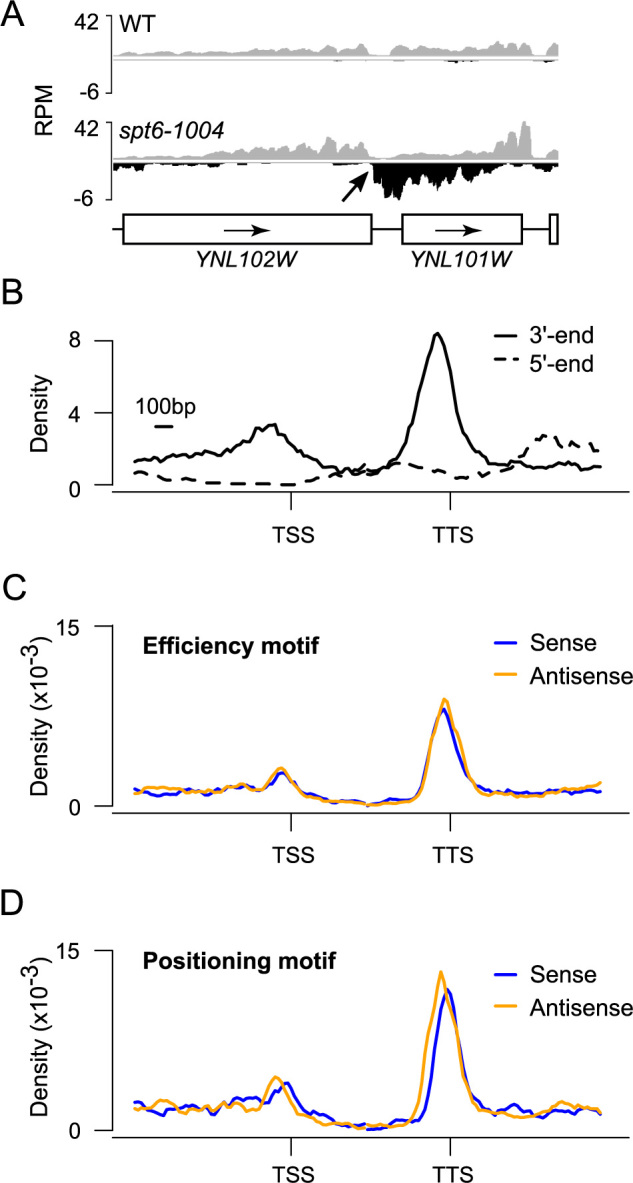
Antisense cryptic transcripts are polyadenylated and tend to terminate at the 3΄-end of adjacent genes. (A) A genome-browser snapshot illustrating a terminator (YNL102W) efficiently terminating an antisense cryptic transcript. RNA-Seq signal on the Watson (gray) and Crick (black) strands is shown. (B) Aggregate profile of antisense starting (dashed trace) and ending (solid trace) positions over genes. (C) Aggregate profile of the efficiency motif (UAUAUA, UACAUA, UAUGUA) enrichment on the sense (blue) and antisense (gold) strands over genes. (D) Aggregate profile of the positioning motif (AAUAAA, AAAAAA) enrichment on the sense (blue) and antisense (gold) strands over genes. For panels ‘B’, ‘C’ and ‘D’ the analyses were done over 1408 yeast genes that are oriented in a divergent, tandem manner (← → →) such that the gene upstream of the TSS is transcribed right to left, while the gene to the right of the TTS is transcribed left to right. This insures that terminators of adjacent genes are not adjacent to the gene of interest.
