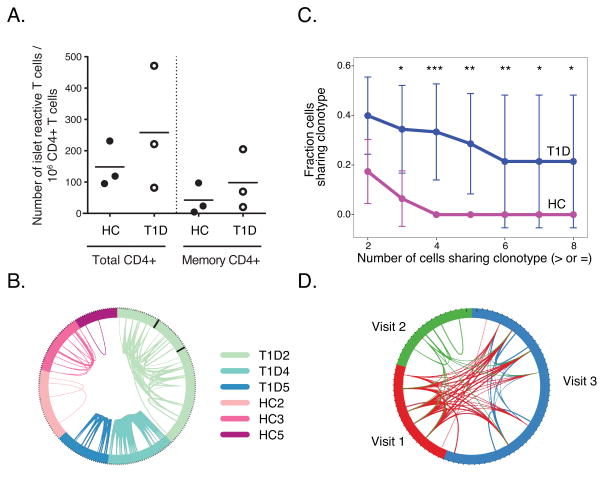
Figure 3. Sharing of rearranged TCRs from islet- antigen reactive CD4+ memory T cells.
A) Levels of islet- antigen reactive CD4+ memory T cells in T1D and HC subjects studied. Cell frequency per million CD4+ T cells was calculated as E/(Tx50) where E is the number of CD4+CD154+CD69+ T cells (Total CD4+) or CD4+CD154+CD69+CD45RA-RO+ (memory CD4+) following enrichment, and T is the total number of CD4+ T cells in 1/50th of the sample pre-enrichment as determined by flow cytometry (p-values ≥0.35, one-sided Wilcoxon test. Symbols represent individual subjects and the bars indicate the mean islet T cell frequency for the subjects in a column. B) TCR sharing in individual islet- antigen reactive T cells. Shown is a circos plot where segments in the circle represent individual cells yielding a rearranged TCR sequence. Black lines for subject T1D2 separate cells from different visits. Arcs connect cells sharing identically rearranged TCR genes. Line thickness is proportional to the number of junctions shared between cells, generally indicating that both TRAV and TRBV junctions were identified. Libraries from three different visits for subject T1D2 were combined for this and subsequent analyses (N = 22, 19 and 52 libraries for visits 1–3, respectively). C) Fraction of cells with expanded clonotypes is higher in T1D than HC cells. Shown are mean fractions of cells ± SD (Y axis) sharing clonotypes with different numbers of cells (X axis). Significance of mean differences between groups was calculated by permutation testing (Materials and Methods) (*, p-value <0.05 and ≥0.01; **, p-value <0.01 and ≥0.001; ***, p-value <0.001). D) Sharing of rearranged TCR junctions over time in subject T1D2. The circos plot depicts each visit in a different color and segments represent individual cells yielding a rearranged TCR sequence at a given visit. Lines connect cells sharing clonotypes at the same or different visits. Experiments were performed on cells from 3 healthy individuals and 3 T1D patients. 92, 35, and 28 cells were analyzed from T1D2, T1D4, and T1D5, respectively. 37, 31, and 22 cells were analyzed from HC2, HC3, and HC4 respectively.