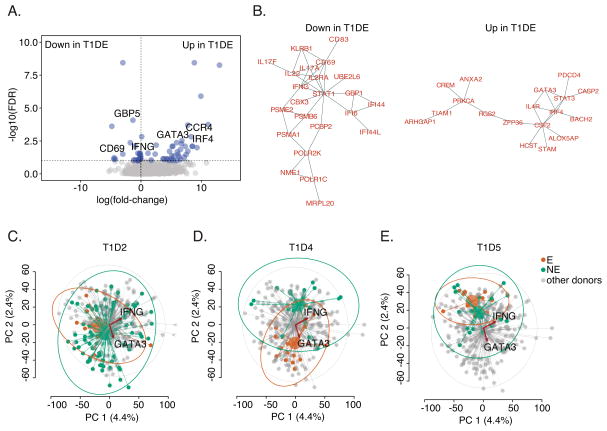
Figure 7. Differential gene expression by expanded clones of islet- antigen reactive CD4+ cells.
A) Genes differentially expressed in islet- antigen reactive CD4+ memory T cells with the most expanded TCRs was determined using a model comprising terms for TCR clonotype frequency and cellular detection rate (31) (https://github.com/linsleyp/Cerosaletti_Linsley). Blue dots, genes differentially expressed with FDR <0.10 (N=62); grey dots, all other genes (N = 5,259 total). Selected immune genes are labeled. Up in T1D-E, genes higher in cells having the most expanded TCRs (including GATA3, CCR4, and IRF4); Down in T1D-E, genes lower in cells having the most expanded TCRs (including IFNG, CD69, and GBP5). B) Significantly interconnected (FDR ≤1.8e-3) PPI networks (32) were found in differentially expressed genes that are preferentially associated with TCR clonotype frequency as defined in (A) (31). Similar interactions were seen in other PPI networks (33). C–E) PCA plots showing PC1 versus PC2 for single cell transcript profiles from islet- antigen reactive CD4+ memory T cells from individual T1D subjects. In each panel, cells from a specified subject are highlighted. Small dots represent individual cells; large dots, centroids for each group; lines, connections between individual cells and centroids; and ellipses, 95% confidence intervals for each group. T1D-E, cells from the specified subject having a rearranged TCR shared with ≥4 cells (orange); T1D-NE, cells from the specified subject having a rearranged TCR shared with <4 cells (green); or others, cells from all other subjects (grey). Biplot vectors show information on expression of individual genes, IFNG and GATA3. Plots summarize the results from 3 T1D patients. 92, 35, and 28 cells were analyzed from T1D2, T1D4, and T1D5, respectively.