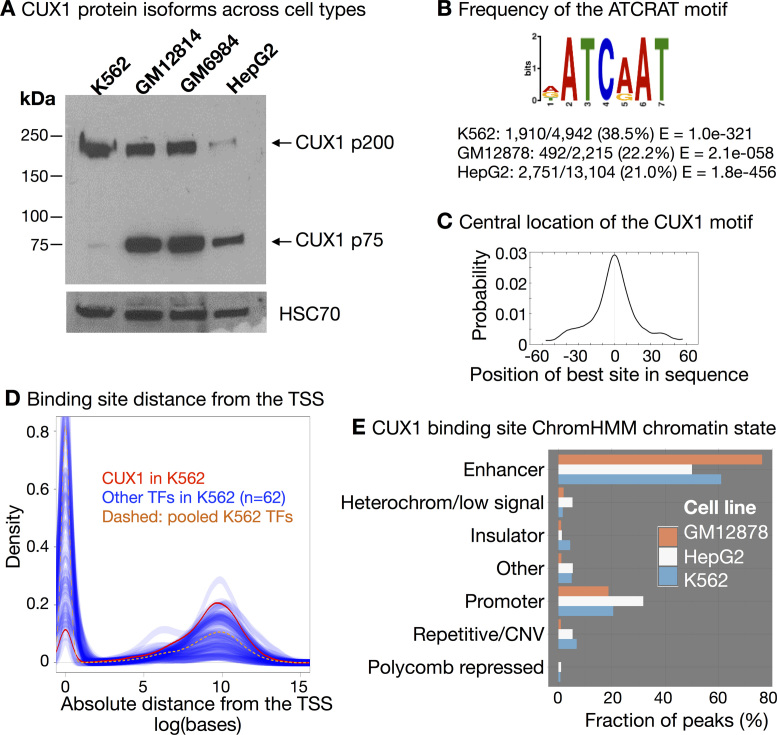
Figure 1.
CUX1 binds to distal enhancers across three cell types. (A) Western blot of CUX1 in human cell lines from three cell types: myeloid leukemia (K562), lymphoblastoid (GM12814 and GM6984) and liver (HepG2). (B) The ATCRAT DNA binding motif is enriched in CUX1 binding sites. The position weight matrix from MEME-ChIP (46) analysis of K562 peaks (IDR < 0.05) is shown. The position weight matrix was similar in the other cell types. The frequency of the ATCRAT DNA binding motif within CUX1 peaks across cell types is indicated. (C) The ATCRAT motif is enriched in the center of CUX1 binding sites. The graph shows the probability of an ATCRAT motif for each base pair upstream and downstream of the center of K562 CUX1 peaks (P < 4.1e–38). The graph was similar in the other cell types. (D) Graph shows the density of CUX1 sites (in red) in relationship to the absolute distance from the single nearest protein-coding genes’ transcription start site (TSS). Blue lines represent the same densities calculated for 62 other ENCODE transcription factor (TF) datasets in K562. The yellow dashed line represents all 62 TFs pooled together. TSSs were defined by Gencode V.24 annotation. Data are representative of other cell types (shown in Supplemental Figure S1). (E) CUX1 is enriched in predicted enhancer chromatin states. Chromatin states are derived from hidden Markov analysis of eight chromatin marks and CTCF ChIP-seq data in each cell type (31). The height of each bar indicates the proportion of peaks in each cell type that fall within the predicted chromatin state for the respective cell type.