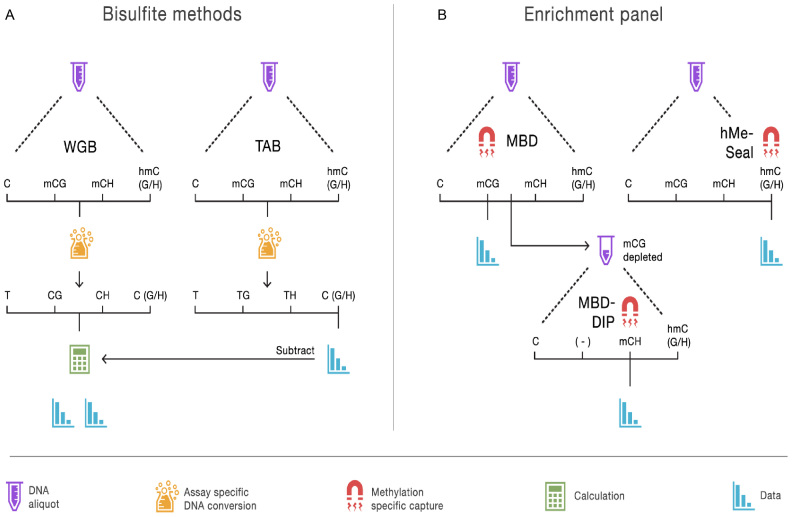
Figure 1.
Overview of methods to assay the brain methylome. (A) The bisulfite methods each start with separate aliquots of genomic DNA. In the case of whole genome bisulfite (WGB) sequencing, sodium bisulfite treatment prior to sequencing converts non-methylated cytosines to uracil but leaves methylated cytosines intact. This provides an estimation of methylation based on the number of unconverted cytosines sequenced. TET-assisted bisulfite (TAB) sequencing sample preparation leaves only hydroxymethylated sites unconverted and provides a direct estimate of hydroxymethylation (hmC). WGB does not discriminate between mC and hmC. However, by subtracting the TAB hmC estimates from the WBS data we obtain estimates of mC. Further distinction between (h)mCG and (h)mCH is achieved by examining the sequence context of each site. (B) With our enrichment panel, genomic DNA is fragmented and fragments are then captured by proteins or other molecules with high affinity for methylated DNA. Next, the non-methylated genomic fraction is removed and the methylation-enriched fraction sequenced. Different sample preparations will enrich for different forms of methylation. Our panel assays mCG using a methyl-CG binding domain (MBD) protein. The MBD protein strictly captures fragments that have methylated CG sites. To assay mCH, we used methylated DNA immunoprecipitation (MeDIP) on DNA fragments that were not captured by the MBD protein. We labeled this approach MBD-DIP. While anti-mC antibodies bind both mCG and mCH, pre-depletion of mCG leads predominantly to capture mCH. For hmC we used a separate aliquot of the genomic DNA, and used selective chemical labeling and capture (hMe-Seal) approach.