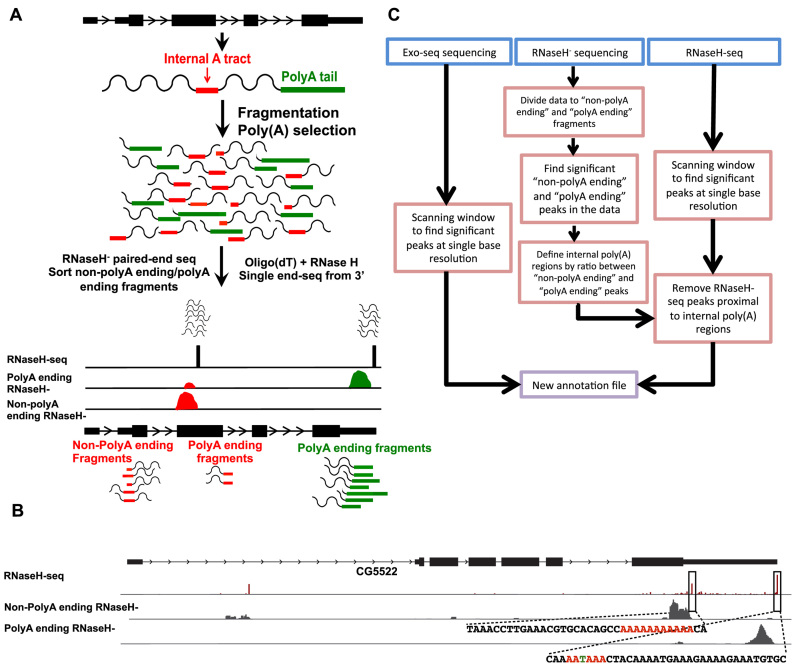
Figure 2.
Experimental procedure for identifying internal poly(A) tracts. (A) Schematic representation of our strategy. RNaseH− reads are sorted to ‘non-polyA ending’ or ‘polyA ending’ reads based on the last few bases of the 3΄ fragments. Then, each group of reads is aligned to the genome separately. A genomic site is classified as a peak of ‘non-polyA ending’ reads based on the ratio of ‘non-polyA ending’ to ‘polyA ending’ aligned reads. (B) An Integrative Genome Viewer plot of the CG5522 gene. Upper track—RNaseH-seq aligned reads. Middle track—reads from RNaseH− libraries not ending with a poly(A) sequence (‘non-polyA ending’ reads). Lower track—reads from RNaseH− libraries ending with a poly(A) sequence (‘polyA ending’ reads). Sequences for the regions surrounding a real gene-end and an internal poly(A) are shown. Colored bases indicate key features: the internal poly(A) tract and the canonical cleavage and polyadenylation site (PAS). (C) Summary of the computational pipeline utilized for generating the new annotation incorporating the newly characterized 5΄ and 3΄ mRNA ends.