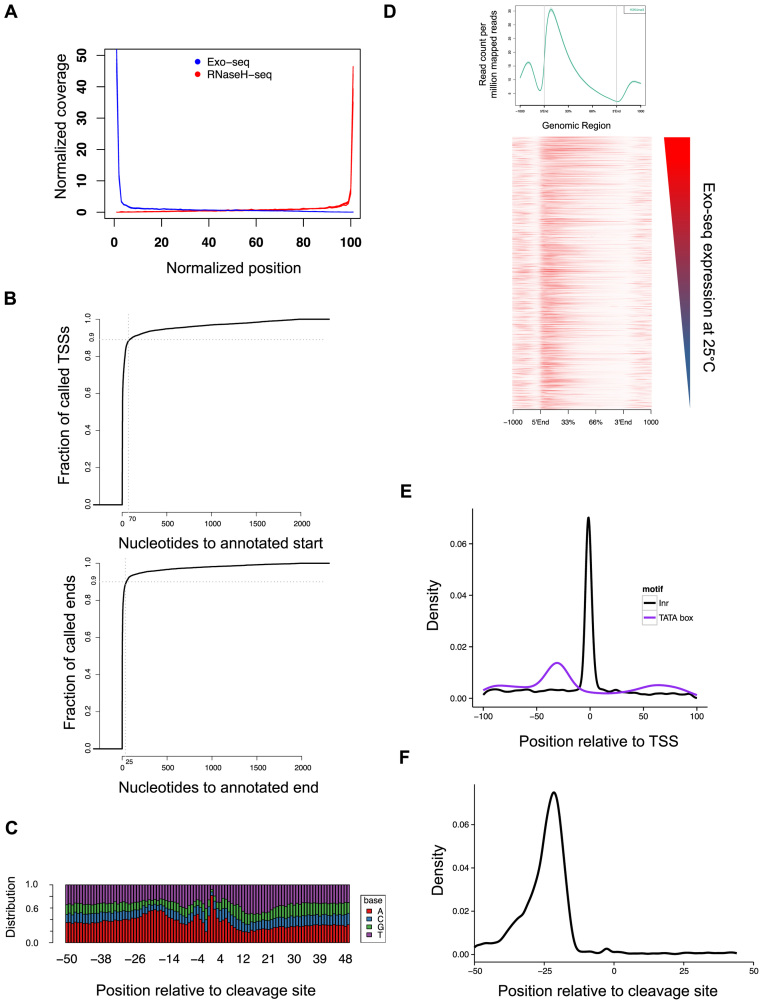
Figure 3.
Validation of the Exo-seq and RNaseH-seq methods. (A) Exo-seq and RNaseH-seq show enrichment for annotated transcript ends. The graph represents the normalized coverage across all genes. The plot includes data from 12 Exo-seq libraries and 6 RNaseH-seq libraries. (B) Distance of the closest annotation determined by Exo-seq (upper panel) or RNaseH-seq (lower panel) to the published modEncdoe annotation. (C) Base distribution of 3΄-end regions of RNaseH-seq annotated genes that differ from the modEncode annotation in 1–5 bases. (D) Average profile (top) and heatmap of the distribution of H3K4me3 ChIP-seq reads (bottom). Average profile is based on read distribution across all gene annotations, while the heatmap presents the top 5000 highly expressed genes at 25°C, sorted by expression. (E and F) Distribution of the position of known motifs with respect to discovered transcript ends: promoter motifs TATAWA (TATA box) and TCAKT [initiation motif (Inr); E] and the canonical cleavage site AAUAAA (F).