Abstract
The human microbiomes across the body evidently interact with various signals in response to biogeographical physiological conditions. To understand such interactions in detail, we investigated how the salivary microbiome in the oral cavity would be regulated by host-related signals. Here, we show that the microbial abundance and gene participating in keeping the human salivary microbiome exhibit global circadian rhythm. Analysis of the 16S rRNA sequences of salivary microbial samples of six healthy adults collected at 4-h intervals for three days revealed that the microbial genera accounting for 68.4–89.6% of the total abundance were observed to significantly oscillate with the periodicity of ∼24 h. These oscillation patterns showed high variations amongst individuals, and the extent of circadian variations in individuals was generally lower than that of interindividual variations. Of the microbial categories oscillated, those classified by aerobic/anaerobic growth and Gram staining, Firmicutes including Streptococcus and Gemella, and Bacteroidetes including Prevotella showed high association with the circadian oscillation. The circadian oscillation was completely abolished by incubating the saliva in vitro, suggesting that host’s physiological changes mostly contributed to the microbial oscillation. Further metagenomic analysis showed that circadian oscillation enriched the functions of environmental responses such as various transporters and two-component regulatory systems in the evening, and those of metabolisms such as the biosynthesis of vitamins and fatty acids in the morning.
Keywords: salivary microbiome, circadian rhythm, 16S rRNA gene, oral, human
1. Introduction
The salivary microbiome is composed of ∼700 species1 mostly belonging to the five phyla, Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes, and Fusobacteria,2,3 similar to other oral microbiomes in the tongue, throat, and tonsil but relatively distinct from the plaque microbiome.4–6 The salivary microbiomes can be characterized by a significantly lower interindividual variability than microbial communities of other body sites such as the gut,7,8 but it is possible to differentiate individuals by phylogenic approaches.9 Though the salivary microbiome is associated with gut microbiome,10,11 the diversity in salivary microbiomes showed relatively low associations with several affecting factors including long-term diet,12 weight and gender,13 geographic location,14 and antibiotic treatment (high tolerance)15 compared with that of the gut microbiome. These apparent discrepancies between the salivary microbiome in the oral cavity and the gut microbiome in the intestine suggest the existence of different regulations and responses between them.
On the other hand, several studies reported the association of the salivary microbiome with not only oral manifestations such as periodontitis and dental caries,16 Behcet’s syndrome and aphthous stomatitis,17 but also with several systemic diseases including pancreatic cancer,18 inflammatory bowel disease (IBD),19 celiac disease,20 rheumatoid arthritis,21 and hepatic cirrhosis.22 These reports suggest that physiological changes at the distal lesion sites largely contribute to the alteration of salivary microbiome, and that the salivary microbiome appears to be more sensitive to host-related rather than environment-related factors in comparison with the gut microbiome.
In this context, it is conceivable that the oral circadian rhythm as a host-related diurnal signal may affect the salivary microbiome in addition to salivary gland and epithelial homeostasis in the oral cavity.23 There have been several researches on the analysis of changes or stability in longitudinal salivary samples collected daily, monthly, or yearly.8,13,24,25 These studies showed the tendency of high stability of the human salivary microbiome over time with low intra-subject variations. Some of the papers also showed more shared species and lower taxonomic changes among the individual salivary than gut microbiomes.13,24 Unlike the gut microbiome, these observations further suggested that host-related internal signals had more impact on the salivary microbiome than external signals related to individual lifestyles. However, more recently, the individual salivary microbiomes collected with 4-h intervals in a day was reported to show little or no change in the microbial profiles.26
In this study, we examined diurnal changes in salivary microbiomes longitudinally collected from six healthy subjects with 4-h intervals for 3 days by 16S rRNA gene and metagenomic sequencing of the salivary DNA. Our data revealed that the majority of salivary microbes exhibited the circadian rhythm profiles with ∼24-h periodicity.
2. Materials and methods
2.1. Subjects and saliva sample collection
Six healthy volunteers were recruited. All subjects were informed of the purpose of this study. This study was approved by the ethical committees of the University of Tokyo and Azabu University, and written consent was obtained from all subjects. Fresh unstimulated saliva (∼1 ml) were collected every 4 h at 12:00, 16:00, 20:00 in the first day, at 0:00, 4:00, 8:00, 12:00, 16:00, 20:00 in the second and third days, and at 0:00, 4:00, 8:00 in the fourth day, accounting for a total of 18 samples for each subject for 3 days (Supplementary Table S1). All subjects took a sleep from 0:00 to 8:00, and shortly awakened at 4:00 for sampling. The sampling of saliva was carried out at least 2 h after meal or toothbrushing. Collected saliva samples were kept at 4°C, frozen in liquid nitrogen within 24 h after sampling, and stored at -80°C until use for DNA preparation.
2.2. DNA preparation and sequencing
Microbial DNA was prepared from saliva samples according to the method described previously.19 The 16S rRNA gene hypervariable V1–V2 regions were PCR-amplified using universal primers (a barcoded 27Fmod and 338R) and, multiplexed amplicon pyrosequencing was carried out using 454 GS FLX Titanium or 454 GS JUNIOR (Roche Applied Science).
To obtain metagenomic sequences, we performed the whole-genome shotgun sequencing with the MiSeq (2 × 300 paired-end run) and IonProton (200-base reads with the Proton I chip) platforms according to the manufacturer’s protocol. Statistics of the sequencing was summarized in Supplementary Tables S1 and S2, respectively.
2.3. Quantitative PCR (qPCR)
The universal bacterial 16S rRNA primers, 27Fmod (5ʹ-agrgtttgatymtggctcag-3ʹ) and 338R (5ʹ-tgctgcctcccgtaggagt-3ʹ) were used to estimate the microbial cell number by real-time qPCR using Escherichiacoli 16S rRNA gene sequence as a standard.
2.4. Data processing of 16S sequences
The 16S V1–V2 sequences were assigned to samples on the basis of their barcode sequence. Reads with an average quality value (QV) < 25 and those lacking the primer sequences at both ends were filtered off. Possibly chimeric sequences that had alignment lengths of < 90% coverage with authentic reference 16S sequences in the database (described below) were also removed. Filter-passed reads obtained from these quality-check processes accounted for ∼50% of the total reads. The majority of the removed reads represented those lacking PCR primer sequences. After trimming both primer sequences from the filter-passed reads, 3,000 reads per sample were randomly selected and used for further analyses.27 The high-quality reads were sorted according to the QV19 and then clustered into operational taxonomic units (OTUs) using a 96% pairwise-identity cutoff with the UCLUST program v.5.2.32 (http://drive5.com/usearch/manual/uclust_algo.html (31 January 2017, date last accessed)). The representative sequences of the generated OTUs were blasted to our own 16S database constructed in this study by GLSEARCH program to determine the closest taxa. The 16S database was constructed from three publically available databases: Ribosomal Database Project (RDP) v.10.31, CORE (http://microbiome.osu.edu/(31 January 2017, date last accessed)), and the reference genome sequence database obtained from the NCBI FTP site (ftp://ftp.ncbi.nih.gov/genbank/ (December 2011, date last accessed)). For assignment of OTUs to the taxa at the phylum, genus and species level, sequence similarity thresholds of 70, 94, and 96% were applied. The results of the taxonomic assignment of the OTUs were summarized in Supplementary Table S3. UniFrac distance28,29 was calculated to assess the dissimilarity (distance) between each sample.
2.5. Data processing of metagenomic sequences
In Ion Proton reads, artificially duplicated sequences were removed using replicate filter if any sequences have a similarity of ≥95% identity to other sequences with identical starting position. Human-derived reads were removed by mapping them to the human genome (HG19) with Newbler. Finally, low-quality reads with average QV < 20 or < 75 bp were removed. For MiSeq reads, low quality bases (QV < 20) at the 5ʹ end of reads were trimmed. Reads in which more than half of bases are QV < 20 and reads of which the length was < 50 bp were filtered out. These procedures were done using FASTX-Tool kit. The filter-passed reads were mapped to the human genome using bowtie2 and the mapped reads were also removed. Reads derived from phiX were also mapped to the phiX genome and the mapped reads were removed.
The high-quality reads were then assembled into contigs by using the Newbler algorithm (v.1.1.03, provided with the GS FLX sequencer). Gene prediction was performed using MetaGeneAnnotator30 with the –m option for multiple species. The nucleotide sequences of predicted genes were clustered using cd-hit-est v.4.6 with a ≥95% sequence identity and ≥90% coverage. The clustered gene sequences were then translated to proteins using the NCBI translation table 11. Functional annotation of the predicted proteins was performed using BLASTP v.2.2.25 against KEGG database with an E-value 1e-5. We mapped 100 K high-quality reads onto the non-redundant gene set using bowtie2 to obtain the quantified functional composition in each sample. Also, we quantitatively evaluated the microbial composition by mapping 100 K high-quality reads onto the reference genomes under the conditions described previously.31
2.6. Periodicity discrimination method (PDM)
In this study, we developed a PDM for evaluation of the periodicity of oscillation patterns of the relative abundance of given taxa and microbial categories.
The microbial relative abundance data at sampling times in a day (12:00, 16:00, 20:00, 0:00, 4:00, 8:00) was normalized as follows.
dn denotes each value of the day, dnorm denotes the normalized value, dmin denotes the minimum value of the day and dmax denotes the maximum value of the day. Normalized values of each day were all in the interval [0, 1].
To figure out the periodicity of a given taxon X in a subject, the normalized abundance data of every 4 h for 3 days were prepared as a list and the average normalized abundance of each time zone was calculated as shown below.
| 12:00 | 16:00 | 20:00 | 0:00 | 4:00 | 8:00 | |
|---|---|---|---|---|---|---|
| Day1 | x 11 | x 12 | x 13 | x 14 | x 15 | x 16 |
| Day2 | x 21 | x 22 | x 23 | x 24 | x 25 | x 26 |
| Day3 | x 31 | x 32 | x 33 | x 34 | x 35 | x 36 |
| Average | X 1 | X 2 | X 3 | X 4 | X 5 | X 6 |
Then, the list was sorted with time zones (column vector in the shown table) by the average normalized abundance of each time zone. For example, when the average normalized abundance lined as follows; X2 > X4 > X5 > X3 > X1 > X6, the sorted list would be lined as shown below.
| TimeZone1 16:00 | TimeZone2 0:00 | TimeZone3 4:00 | TimeZone4 20:00 | TimeZone5 12:00 | TimeZone6 8:00 | |
|---|---|---|---|---|---|---|
| Day1 | x 12 | x 14 | x 15 | x 13 | x 11 | x 16 |
| Day2 | x 22 | x 24 | x 25 | x 23 | x 21 | x 26 |
| Day3 | x 32 | x 34 | x 35 | x 33 | x 31 | x 36 |
| Average | X 2 | X 4 | X 5 | X 3 | X 1 | X 6 |
This sorting operation is especially useful to reduce the calculation resources when the observed time points are further large, so that combination explosion can be avoided for selecting all possible combination of TimeZones.
Then, sorted time zones in the sorted list are grouped as a combination of two separated ‘TimeZoneCombi’ which discern the deviation of the abundance between the two time zones. By using the following algorithm, the best P-value for a taxon and the TimeZoneCombi can be obtained.
BestPvalue ← 1
FOR each TimeZoneCombi
BestPvalueOfTimeZone ← 1
FOR each Threshold = each data of taxa X
Pvalue ← CALL FisherExactTest WITH taxa X data in time zones of TimeZoneCombi, taxa X data of other time zones AND Threshold
IF Pvalue < BestPvalueOfTimeZone THEN
BestPvalueOfTimeZone ← Pvalue
BestThresholdOfTimeZone ← Threshold
END IF
END FOR
IF BestPvalueOfTimeZone < BestPvalue THEN
BestPvalue ← BestPvalueOfTimeZone
BestThreshold ← BestThresholdOfTimeZone
BestTimeZone ← TimeZoneCombi
END IF
END FOR
In the subprocedure ‘FisherExactTest’, the P value from the test is computed according to the 2 × 2 contingency table shown below.
| ≥ Threshold | < Threshold | |
|---|---|---|
| TimeZoneCombi | a | b |
| Other time zone | c | d |
a: the number of time points of taxon X data in time zones of TimeZoneCombi which is equal or over the threshold.
b: the number of time points of taxon X data in time zones of TimeZoneCombi which is under the threshold.
c: the number of time points of taxon X data in other time zones which is equal or over the threshold.
d: the number of time points of taxon X data in other time zones which is under the threshold.
Thus, the less P value the taxon has, the stronger periodicity it is regarded to have.
Furthermore, the time zones determined by TimeZoneCombi with the best P value are regarded as proliferating time zones for the taxon X.
The whole procedures were depicted in Supplementary Fig. S1.
3. Results
3.1. Circadian rhythm in the salivary microbiome
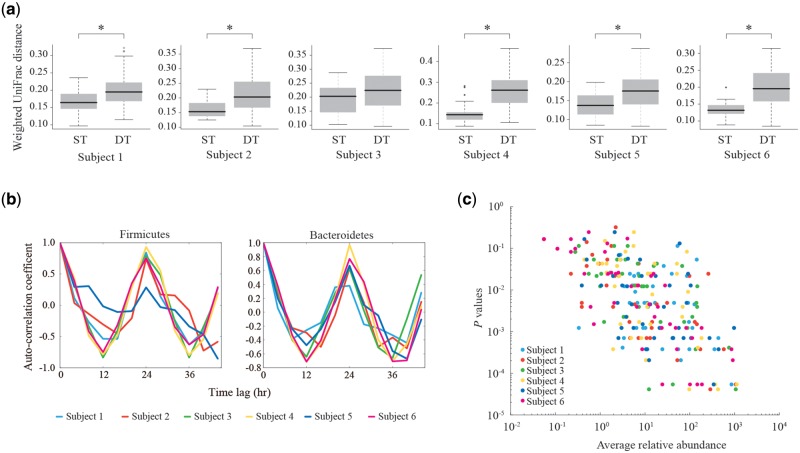
We longitudinally collected a total of 108 unstimulated salivary samples (18 samples per subject) every 4 h for 3 days from six healthy subjects (two females and four males). We enumerated sequences of the 16S rRNA gene V1–V2 region by pyrosequencing the salivary DNA with the Roche 454 platform (see Materials and methods). The 16S reads were clustered to obtain the OTUs for each subject, and the generated OTUs were assigned to taxa by blasting against the 16S databases (Supplementary Table S3). The phylum- and genus-level assignment apparently showed the diurnal change in the microbial abundance for all the subjects (Supplementary Fig. S2). We next performed the UniFrac distance analysis of the microbial communities, in which the distances between the samples at the same times (ST) and those at different times (DT) for 3 days were measured, respectively. The results revealed that the UniFrac distances between the ST samples were lower than those between the DT samples for all the subjects, of which the five subjects showed a statistical significance, indicating that the overall structure of the ST samples was more similar than that of the DT samples (Fig. 1a).
Figure 1.
Detection of diurnal oscillation in salivary microbiomes. (a) Comparison of the weighted UniFrac distances between the ST and DT samples in each subject. Asterisks indicate the statistical significance (P < 0.05, Wilcoxon signed-rank test). (b) Autocorrelation coefficients of the relative abundance of Firmicutes, Bacteroidetes in all the subjects. The horizontal axis represents time lag used for autocorrelation calculation. (c) Correlations between the relative abundance and circadian periodicity of the genera. Genera with ≥0.1% average relative abundance in all the subjects were analysed. The vertical axis represents raw P values of the circadian periodicity of each genus calculated by the PDM. The horizontal axis represents the average relative abundance of each genus.
We then performed the autocorrelation analysis for diurnal changes in the relative abundance of the two major phyla, Firmicutes and Bacteroidetes in each subject between the 3 days. The analysis showed that the highest correlation coefficient was observed at ∼24-h interval in all of the subjects (Fig. 1b). These data strongly suggested that the observed diurnal changes in the microbial abundance of the salivary microbiomes exhibited a circadian rhythm at the phylum level. Similar results were also obtained from the analysis using the microbial abundance data estimated from mapping of the metagenomic reads on the reference genomes (Supplementary Fig. S3).
On the other hand, the hierarchical clustering of the samples based on the Spearman's rank correlation coefficient revealed that many of the ST samples but not all were clustered most closely within a subject, whereas all the samples were clearly aggregated by the individuals (Supplementary Fig. S4), suggesting that the extent of circadian intra-subject variations was generally lower than that of the inter-subject variations.
To detect the taxa with a statistically significant circadian periodicity, we developed the PDM that determines the P value for the circadian periodicity of each genus by using the Fisher’s exact test repeatedly for every possible threshold and time-zone combination (Supplementary Fig. S1). The PDM analysis followed by Bonferroni correction of 19 genera (18 known and one undefined) with ≥ 0.1% relative abundance in all of the six subjects found that the genera accounting for 68.4–89.6% of the total abundance (∼79.3% in average for the six subjects) exhibited a significant oscillation with the 24-h periodicity (P < 0.05), among which Prevotella, Gemella, Peptostreptococcus, Solobacterium, Streptococcus, and Haemophilus were statistically significant for all the subjects (Supplementary Table S4). Furthermore, we found a correlation between the relative abundance of the genera and the P value of the circadian periodicity in toto, which indicated that the higher abundant genera tended to have the stronger circadian periodicity, and vice versa (Fig. 1c). For the major four phyla, the PDM analysis also indicated the high circadian periodicity of them in most of the subjects as well as the autocorrelation analysis mentioned above (Supplementary Table S5). We also evaluated the circadian periodicity of the genera and phyla by JTK_CYCLE32 (Supplementary Tables S4 and S5). In the genus level, JTK_CYCLE and PDM both showed the significant periodicity in Prevotella for the five subjects, Neisseria for the four subjects, Gemella and Streptococcus for the three subjects. In the phylum level, Bacteroidetes for the five subjects, Firmicutes for the four subjects, Proteobacteria for the three subjects, and Actinobacteria for the two subjects were significant in the circadian periodicity, respectively.
In addition, we examined the diurnal change in the OTU number. We compared quantitative differences in all pairs of OTU numbers in the ST and those in the DT samples in each subject, but no significant difference was observed between them for all the subjects, suggesting that OTU number representing the species richness had no circadian oscillation (Supplementary Fig. S5).
We further analysed changes in the microbial cell number by qPCR with universal 16S primers. The results observed statistically significant periodicity of microbial cell numbers in two of the six subjects, in which microbial cell number increased at 4:00 and 16:00 in the subject 3, and at 4:00 and 8:00 in the subject 4 (Supplementary Table S6 and Supplementary Fig. S6). These data suggested that microbial cell number also exhibited the circadian oscillation with individual differences.
3.2. Characterization of circadian oscillation patterns
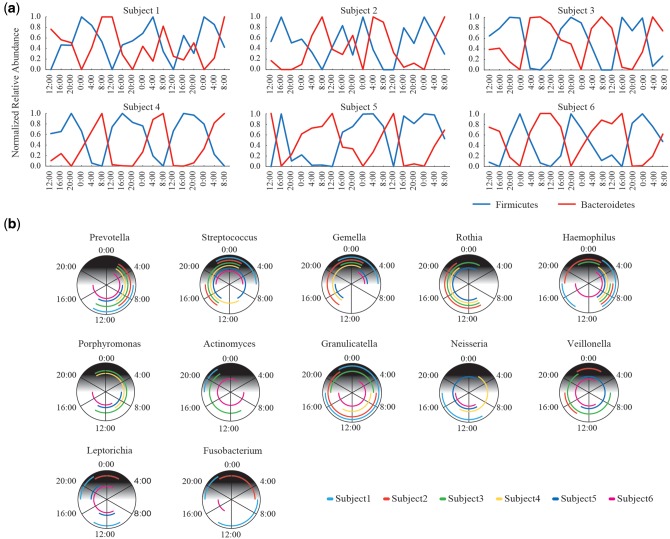
We characterized the oscillation pattern of salivary microbes at the phylum and genus level. We found that the relative abundance of Firmicutes was increased in the evening, whilst that of Bacteroidetes was in the daytime in all the subjects, indicating that these two major phyla had an inverse relation in the circadian oscillation pattern (Fig. 2a). At the genus level, we showed proliferation time zones of the circadian oscillation of the 12 genera with ≥ 1% relative abundance in all the subjects, respectively (Fig. 2b). Among the 12 genera, Prevotella, Gemella, Streptococcus, and Haemophilus showed the significant circadian periodicity for all the subjects, but the other eight genera showed non-significant periodicity in a few subjects (Supplementary Table S4). The relative abundance of Prevotella was increased in the morning (from 4:00 to 12:00) in all the subjects, and those of Gemella and Streptococcus were increased in the evening (from 16:00 to 0:00) in all the subjects. The relative abundance of Haemophilus was proliferated in the morning (around 8:00) in all the subjects, but different proliferation time zones were also observed in three subjects. These genera are typical examples that had almost same circadian oscillation patterns shared among all the six subjects. The inverse oscillation pattern between Firmicutes and Bacteroidetes described above could be largely due to the strong circadian periodicity of the top two pre-dominant Streptococcus and Prevotella genera belonging to these two phyla, respectively. Rothia had similar proliferation time zones in the relative abundance among four subjects but no significant circadian periodicity in two subjects. The other genera such as Granulicatella and Veillonella did not share similar proliferation time zones among the subjects with the significant circadian periodicity. These data revealed that a few but highly abundant genera had the strong circadian periodicity and similar circadian oscillation patterns shared with most of the subjects. Many of the genera had the significant circadian periodicity but their oscillation patterns were considerably varied among the subjects. Thus, these data suggested the high inter-subject variation in the circadian oscillation profile.
Figure 2.
Oscillation patterns of major taxa. (a) Circadian oscillation patterns of Firmicutes and Bacteroidetes. Changes in the normalized relative abundance of Firmicutes and Bacteroidetes in each subject are represented. (b) Circadian proliferation time zones of major genera. The results of the genera with ≥1% relative abundance in all the subjects are shown. Lines cover the proliferation time zones of the genus in each subject determined by PDM, in which the subjects having the proliferation time zones with no significant circadian periodicity were eliminated (see Supplementary Table S4).
3.3. Bacterial phenotypes and oscillation patterns
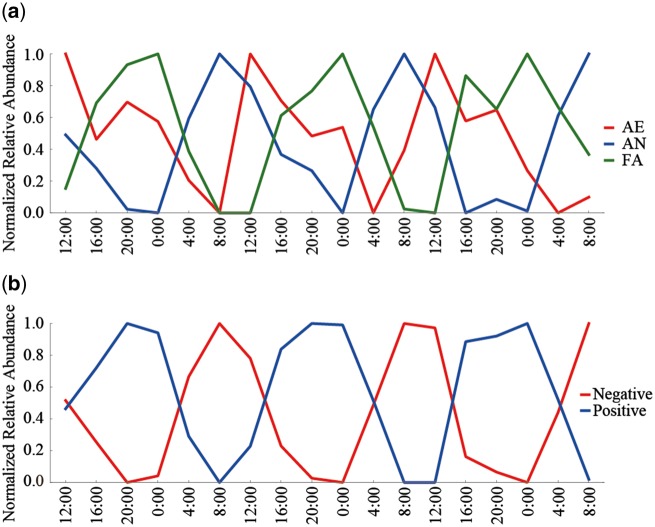
To examine if the oscillation patterns of the salivary microbiome is determined by the bacterial phenotype, we compared the oscillation patterns between strict aerobes (AE), facultative anaerobes (FA), and strict anaerobes (AN). The PDM analysis revealed that the relative abundance of AE, FA, and AN had significant circadian oscillations in all the six subjects, respectively (Supplementary Table S7). We analysed changes in the average relative abundance of the three microbial categories through the three days (Fig. 3a). The results showed differences in the circadian oscillation patterns among the three categories, in which AN was proliferated in the morning between 4:00 and 12:00, and FA was in the evening between 16:00 and 4:00, indicating the circadian oscillation pattern of AN and FA had an inverse relation. On the other hand, the proliferation time zone of AE was the daytime between 8:00 and 20:00, which was intermediate between those of AN and FA. In the 19 genera with ≥0.1% relative abundance in all the subjects, Neisseria, Rothia, and Campylobacter were included in the typical AE pattern, Prevotella and Solobacterium were in the typical AN pattern, and Streptococcus, Actinomyces and Gemella were in the typical FA pattern, respectively (Supplementary Fig. S7). These data revealed that the three categories defined by the bacterial aerobicity were sequentially proliferated in the relative abundance with some overlap among them in a day. The periodicity of the three microbial categories was also evaluated by JTK_CYCLE, showing similar results to those by the PDM (Supplementary Table S7).
Figure 3.
Circadian oscillation patterns of the bacterial phenotypes. (a) Oscillation patterns of genera classified by the bacterial aerobicity. The oscillation patterns of strictly aerobic (AE), facultative anaerobic (FA), and strictly anaerobic (AN) genera are shown. (b) Oscillation patterns of genera classified by the Gram stainability. The oscillation patterns of Gram-positive and -negative genera are shown.
We also compared the oscillation pattern between Gram-positive and -negative species. The PDM and JTK_CYCLE analyses revealed that the relative abundance of Gram-positive and -negative genera had significant circadian oscillations in all the subjects, respectively (Supplementary Table S8). The analysis indicated that the Gram-positive species was proliferated in the evening between 16:00 and 4:00, whereas that of the Gram-negative species was in the day between 4:00 and 16:00, respectively (Fig. 3b). In the 19 genera with ≥0.1% relative abundance, oscillation patterns of several Gram-negative Prevotella and Haemophilus were similar to that of the all Gram-negative species, and those of several Gram-positive Streptococcus, Rothia, Actinomyces and Gemella were similar to that of the all Gram-positive species (Supplementary Fig. S8). Furthermore, the oscillation pattern of Gram-positive bacteria shown in Fig. 3b appeared to be overlapped with those of AE and FA shown in Fig. 3a, whereas that of Gram-negative bacteria appeared to be overlapped with those of AE and AN. The proportion of genera classified by the Gram stainability and aerobicity in samples of the subjects was shown in Supplementary Table S9.
3.4. Disappearance of circadian rhythm in salivary microbiomes in vitro
Salivary samples from two subjects (subjects 1 and 2) were incubated at 37°C in vitro under aerobic conditions, and changes in the microbial abundance in aliquot samples collected every 1 h for 7 h were monitored by 16S rRNA gene sequence analysis. These in vitro data were compared with those of salivary samples freshly collected with the ST intervals as those in vitro from the same two subjects (Supplementary Fig. S9). The results indicated that the in vitro samples had no longer change in the microbial composition with very low UniFrac distances between them, whilst freshly collected samples showed the circadian oscillation as expected.
3.5. Functional analysis based on metagenomic data
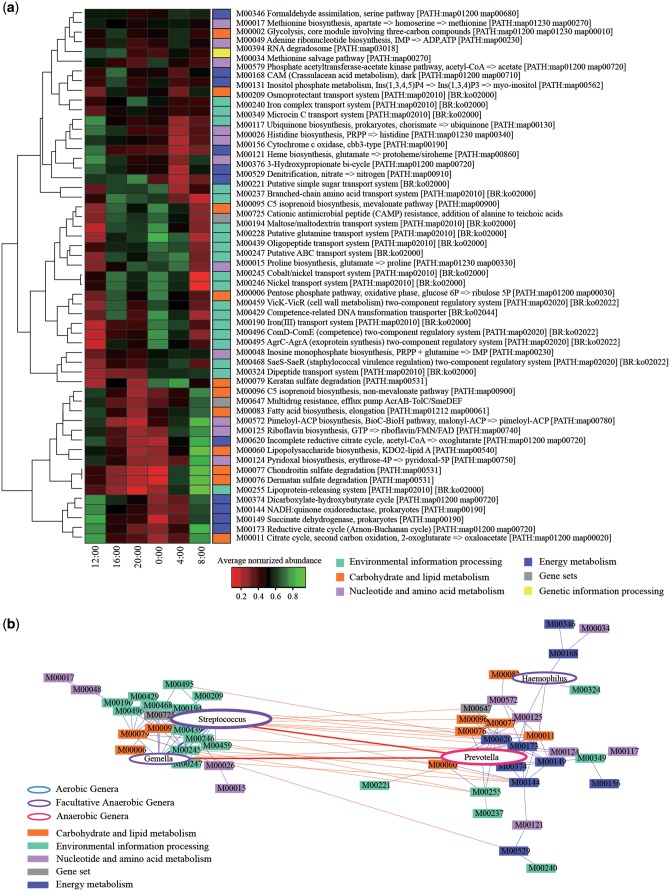
For addressing the biological periodicity, we performed the functional assignment of the annotated genes in the assembled metagenomic sequences, and found a total of 370 KEGG modules by using the KEGG database. We then quantified the 370 KEGG modules by mapping 100 K metagenomic reads per sample to them (see Materials and methods). The results detected 55 modules that significantly exhibited 24 h oscillation in more than five subjects (Fig. 4a). These mainly included the modules assigned to four functional categories, ‘Environmental information processing’, ‘Energy metabolism’, ‘Carbohydrate and lipid metabolism’, and ‘Nucleotide and amino acid metabolism’. Many of the modules assigned to ‘Environmental information processing’ were enriched in the evening (between 16:00 and 4:00), whereas those assigned to the other three functional categories were enriched in the morning (between 4:00 and 12:00), respectively, indicating the inverse relation between ‘Environmental information processing’ and the other three functional categories (Supplementary Fig. S10). The modules assigned to ‘Environmental information processing’ included the functions related to various transporters including those for metal ions and two-component regulatory systems. On the other hand, the modules assigned to ‘Carbohydrate and lipid metabolism’, ‘Energy metabolism’, and ‘Nucleotide and amino acid metabolism’ included the functions for the biosynthesis of vitamins and fatty acids, polysaccharides degradation, Hem biosynthesis from glutamate, the reductive citrate cycle, and electron transport.
Figure 4.
The KEGG modules having the circadian oscillation. (a) Heatmap of the relative gene abundances of 55 KEGG modules. Colour gradient indicate the degree of the average abundance of the KEGG modules which had statistically significance in at least five subjects. Colours on right side indicate six upper functional categories of the KEGG modules. (b) Network of the KEGG modules and genera. The results for the significant correlation between the 55 KEGG modules and four genera in more than five subjects are shown. Interactions between nodes were calculated by Spearman’s rank correlation and correlations with P >0.05 or P <0.6 are removed. Coloured rectangles correspond to the KEGG modules and ellipse nodes signify the genera. Size of ellipse nodes corresponds to the relative abundance of the genera. Blue and red lines indicate positive and negative correlation, respectively.
We further performed the network analysis of the correlation between the 55 modules and the five genera (Streptococcus, Prevotella, Haemophilus, Gemella, and Granulicatella) with ≥1% relative abundance and the significant circadian periodicity shared with more than five subjects (Supplementary Table S4). The analysis formed two distinct negatively correlated clusters, which were represented by positive correlations between Streptococcus and Gemella and many of the modules assigned to ‘Environmental information processing’, and those between Prevotella and Haemophilus and the modules assigned to ‘Energy metabolism’ (Fig. 4b). Granulicatella had no significant correlation with any of the module. These correlations were also related with the bacterial phenotypes, in which Streptococcus and Gemella were the Gram-positive, and Prevotella and Haemophilus were Gram-negative.
4. Discussion
In this study, we demonstrated that the majority of the salivary microbes exhibited the circadian oscillation with ∼24-h in the relative abundance by the analysis using the 16S V1–V2 sequences obtained from the samples collected with 4-h intervals for three consecutive days from six healthy adults. We obtained the several pieces of evidence for the circadian oscillation from significant differences in the UniFrac distances between the samples collected at the same and DT in each subject, and apparent diurnal changes in the relative abundance of taxa at the phylum and genus level with the highest autocorrelation coefficient of ∼24-h interval. Furthermore, the circadian periodicity of each taxon was also statistically evaluated by the PMD originally developed in this study. The basis of the PDM is to assess the reproducibility of proliferation time patterns of the relative abundance of each taxon by the Fisher exact test, so that the PDM is also applicable to the relatively minor taxa of which the 16S read number is zero in several samples. Therefore, the PDM generally enables us to detect more taxa with the periodic pattern than the JTK_CYCLE.32 The PDM analysis found that the major genera had the tendency of relatively high circadian periodicity compared with minor genera in the salivary microbiome, similar to the observation in the gut microbiome.33 However, on the contrary, the recent study reported almost no significant diurnal variation in the salivary microbiome by using the clustering analysis of two sample sets collected every 4 h in 24 h with a 1 week interval from five subjects.26 We similarly performed the hierarchical clustering of the same sample sets, but could not clearly show the closest relation between the samples collected at the ST by this clustering. Therefore, this inconsistency suggested that the hierarchical clustering was insufficient to differentiate subtle changes in variations within individuals.
The circadian oscillation completely disappeared with very low UniFrac distances between the samples in the in vitro incubation experiments, suggesting that the circadian oscillation in the salivary microbiome is not an autonomous process, but a response linked to certain host’s signals and/or oral environmental changes associated with them. The salivary microbiome is thought to be formed by microbes adhered to various intraoral surfaces because the saliva is sterilely secreted from the salivary glands into the oral cavity.3 This implies that the microbes associated with intraoral surfaces may change their releasing level from the surfaces to the saliva and/or their growth rate in the saliva. The microbes in the throat, tonsil, and tongue may be the resources of the salivary microbiome because the salivary microbiome composition is similar to that of these microbiomes.4–6 In this context, the recent paper showed that the plaque microbiome exhibited little circadian oscillation,34 suggesting that the sensitivity to stimuli in the oral cavity differed between the plaque and salivary and possibly tongue and tonsil microbiomes.
We observed differences in the circadian periodicities (P values estimated by the PDM) among various microbial categories by comparing the average P values of 28 microbial categories classified by the aerobicity, Gram stainability, and taxonomically (Supplementary Table S10). The data revealed that the first ten categories had much higher circadian periodicity than others, which included the three categories by the aerobicity, two by the Gram stainability, two phyla (Firmicutes and Bacteroidetes), and three genera (Streptococcus, Prevotella, and Gemella). These oscillation patterns were clearer than those of the others. These data suggested large associations of these microbial categories to the circadian oscillation of the salivary microbiome. On the other hand, the other 18 genera exhibiting less periodicity than the 10 microbial categories had less similarity of the oscillation patterns among most of the subjects, suggesting the high inter-subject variation of these microbes in the circadian regulation of salivary microbiome.
Regarding the bacterial phenotypes, our data suggested that the aerobicity in the oral cavity was also diurnally changed, and more aerobic around noon in a day according to the oscillation pattern of the three microbial categories classified by the aerobicity. In addition, the Gram stainability was also largely involved in the circadian oscillation in the salivary microbiome, in which Gram-positive and -negative species had the trade-off relation of circadian oscillation patterns. At present, we have no reasonable interpretation for the relationship between circadian oscillation and the Gram stainability.
The circadian oscillation of gene functions was also evaluated by metagenomic data. In the daytime, the functions assigned to ‘Energy metabolism’ and ‘Carbohydrate and lipid metabolism’ mainly associated with Prevotella were enriched, and in the evening, those to ‘Environmental information processing’ mainly associated with Streptococcus were enriched, respectively. These findings suggest that functions of salivary microbes devote to environmental responses in the daytime, whereas those devote to metabolisms in the evening, and these functional oscillations may be related with maintenance of the oral homeostasis.
Recently, it was reported that the gut microbiome exhibited the circadian oscillation in the microbial abundance in mice, and the circadian oscillation was abolished by disruption of a host circadian clock gene.33,35 From these observations, it can be suggested that the circadian oscillation of the salivary microbiome is regulated by host clock genes. Although ∼15% of the salivary metabolites were reported to exhibit diurnal oscillation,36 the factors or signals affecting the circadian oscillation of the salivary microbiome are unknown at present and further investigation is needed to address them.
Supplementary Material
Acknowledgements
The authors thank Dr Y. Hirose (Toyohashi University of Technology) for technical support for sequencing, and Drs H. Takayasu (Tokyo Institute of Technology), M. Takayasu (Sony Computer Science Laboratories, Inc.) and T. Yamada (Tokyo Institute of Technology) for their valuable advice in the discussion. The Super-computing resource was provided by the Human Genome Center (The University of Tokyo).
Conflict of interest
None declared.
Accession numbers
DRA005351, DRA005363, DRA5277, DRA3992
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Data availability
All high-quality 16S V1-V2 sequences analyzed in the present study were deposited into the DDBJ/GenBank/EMBL database with accession numbers DRA005351, DRA005363. The filter-passed metagenomic sequences were deposited with accession numbers DRA5277, DRA3992.
Funding
This work was supported in part by a Grant-in-Aid for JSPS Fellows (No. 7826) to L.T., a Grant-in-Aid for Challenging Exploratory Research (No. 25640105) to M.H., and the global COE project of ‘Genome Information Big Bang’ from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan to M.H. and K.O. Funding to pay the Open Access publication charges for this article was provided by an administration grant from the Waseda University.
References
- 1. Aas J.A., Paster B.J., Stokes L.N., Olsen I., Dewhirst F.E.. 2005, Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol., 43, 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zaura E., Keijser B.J.F., Huse S.M., Crielaard W.. 2009, Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol., 9, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takeshita T., Kageyama S., Furuta M., et al. 2016, Bacterial diversity in saliva and oral health-related conditions: the Hisayama Study. Sci. Rep., 6, 22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Segata N., Haake S.K., Mannon P., et al. 2012, Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol., 13, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huttenhower C., Gevers D., Knight R., et al. 2012, Structure, function and diversity of the healthy human microbiome. Nature, 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eren A.M., Borisy G.G., Huse S.M., Welch J.L.M.. 2014, Oligotyping analysis of the human oral microbiome. Proc. Natl. Acad. Sci. USA. 111, E2875–E2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huse S.M., Ye Y.Z., Zhou Y.J., Fodor A.A.. 2012, A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS ONE, 7, e34242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lazarevic V., Whiteson K., Hernandez D., Francois P., Schrenzel J.. 2010, Study of inter- and intra-individual variations in the salivary microbiota. BMC Genomics, 11, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leake S.L., Pagni M., Falquet L., Taroni F., Greub G.. 2016, The salivary microbiome for differentiating individuals: proof of principle. Microbes Infect., 18, 399–405. [DOI] [PubMed] [Google Scholar]
- 10. Ding T., Schloss P.D.. 2014, Dynamics and associations of microbial community types across the human body. Nature, 509, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arimatsu K., Yamada H., Miyazawa H., et al. 2014, Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci. Rep., 4, 04828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Filippis F., Vannini L., La Storia A., et al. 2014, The same microbiota and a potentially discriminant metabolome in the saliva of omnivore, ovo-lacto-vegetarian and Vegan individuals. PLoS ONE, 9, e112373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stahringer S.S., Clemente J.C., Corley R.P., et al. 2012, Nurture trumps nature in a longitudinal survey of salivary bacterial communities in twins from early adolescence to early adulthood. Genome Res., 22, 2146–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nasidze I., Li J., Quinque D., Tang K., Stoneking M.. 2009, Global diversity in the human salivary microbiome. Genome Res., 19, 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zaura E., Brandt B.W., de Mattos M.J.T., et al. 2015, Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. mBio, 6, e01693–e01715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Belstrom D., Fiehn N.E., Nielsen C.H., et al. 2015, Differentiation of salivary bacterial profiles of subjects with periodontitis and dental caries. J. Oral Microbiol., 7, 27429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seoudi N., Bergmeier L.A., Drobniewski F., Paster B., Fortune F.. 2015, The oral mucosal and salivary microbial community of Behcet's syndrome and recurrent aphthous stomatitis. J. Oral Microbiol., 7, 27150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Farrell J.J., Zhang L., Zhou H., et al. 2012, Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut, 61, 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Said H.S., Suda W., Nakagome S., et al. 2014, Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res., 21, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Francavilla R., Ercolini D., Piccolo M., et al. 2014, salivary microbiota and metabolome associated with celiac disease. Appl. Environ. Microbiol., 80, 3416–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X., Zhang D.Y., Jia H.J., et al. 2015, The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med., 21, 895–905. [DOI] [PubMed] [Google Scholar]
- 22. Bajaj J.S., Betrapally N.S., Hylemon P.B., et al. 2015, Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology, 62, 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Papagerakis S., Zheng L., Schnell S., et al. 2014, The circadian clock in oral health and diseases. J. Dent. Res., 93, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. David L.A., Materna A.C., Friedman J., et al. 2014, Host lifestyle affects human microbiota on daily timescales. Genome Biol., 15, R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cameron S.J.S., Huws S.A., Hegarty M.J., Smith D.P.M., Mur L.A.J.. 2015, The human salivary microbiome exhibits temporal stability in bacterial diversity. FEMS Microbiol. Ecol., 91, fiv091. [DOI] [PubMed] [Google Scholar]
- 26. Belstrom D., Holmstrup P., Bardow A., Kokaras A., Fiehn N.E., Paster B.J.. 2016, Temporal stability of the salivary microbiota in oral health. PLoS ONE, 11, e0147472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim S.W., Suda W., Kim S., et al. 2013, Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res., 20, 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lozupone C., Knight R.. 2005, UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol., 71, 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamady M., Lozupone C., Knight R.. 2010, Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J., 4, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Noguchi H., Taniguchi T., Itoh T.. 2008, MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res., 15, 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nishijima S., Suda W., Oshima K., et al. 2016, The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res., 23, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hughes M.E., Hogenesch J.B., Kornacker K.. 2010, JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms, 25, 372–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thaiss C.A., Zeevi D., Levy M., et al. 2014, Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell, 159, 514–529. [DOI] [PubMed] [Google Scholar]
- 34. Sato Y., Yamagishi J., Yamashita R., et al. 2015, Inter-Individual differences in the oral bacteriome are greater than intra-day fluctuations in individuals. PLoS ONE, 10, e0131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liang X., Bushman F.D., FitzGerald G.A.. 2015, Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci. USA, 112, 10479–10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dallmann R., Viola A.U., Tarokh L., Cajochen C., Brown S.A.. 2012, The human circadian metabolome. Proc. Natl. Acad. Sci. USA, 109, 2625–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All high-quality 16S V1-V2 sequences analyzed in the present study were deposited into the DDBJ/GenBank/EMBL database with accession numbers DRA005351, DRA005363. The filter-passed metagenomic sequences were deposited with accession numbers DRA5277, DRA3992.