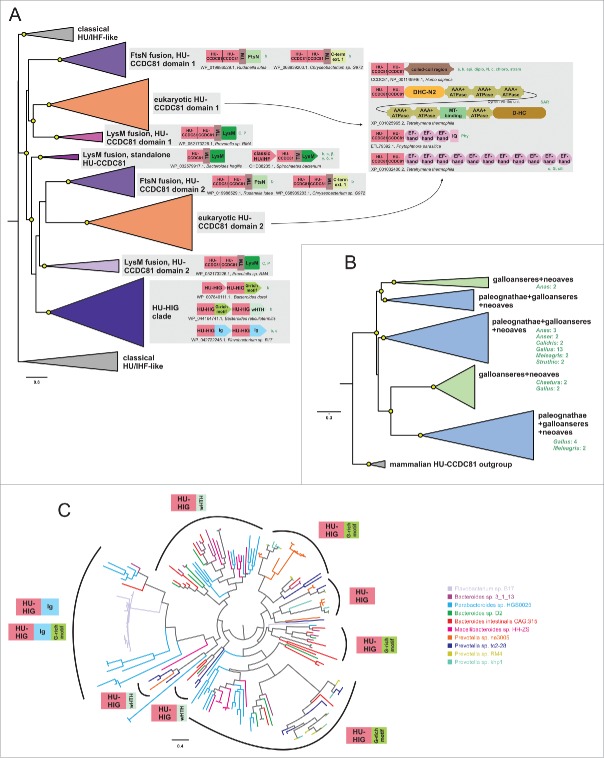
Figure 2.
Phylogenetic relationships and genome associations in HU-CCDC81 and HU-HIG families. (A) Phylogenetic tree depicting higher-level relationships between HU families/clades described in this study. Branches are collapsed at levels containing clearly-delineated monophyletic groups, labeled to the right. Nodes with greater than 65% bootstrap support are marked with yellow circle. Representative conserved domain architectures and gene neighborhoods in a given clade provided to the right. (For complete list see Supplemental Material). Phyletic patterns of a given architecture/neighborhood are found provided to the right in green lettering. Phylogeny abbreviations: b, bacteroidetes; C, Chlamydia; P, Porphyromonas; s, spirochaetes; β, β-proteobacterial; γ, γ-proteobacteria; δ, δ-proteobacteria; v, verrucomicrobia; a, animals; k, kinetoplastids; api, apicomplexa; diplo, diplomonads; N, Naegleria; cili, ciliates; chloro, chlorophytes; stram, stramenopiles; SAR, stramenopile-alveolate-rhizarian group; Phy, Phytopthora; o, oomycetes; G, Guillardia. (B) Phylogenetic tree depicting the multiple paralogs identified in avian expansion of HU-CCDC81 domains. Monophyletic clades, as determined by phyletic distribution conservation patterns, are collapsed and then labeled and colored according to evolutionary depth. Nodes with greater than 70% bootstrap support are marked with yellow circle. Potential lineage-specific expansions within a clade are labeled with total number of non-redundant protein copies and phyletic patterns. (C) Phylogenetic tree depicting rampant LSEs, gene loss, and incomplete lineage-sorting in HU-HIG family based on a set of all HU-HIG sequences retrieved from the 10 bacteroidetes genomes, listed in key to the right, with the highest number of identifiable HU-HIG sequences. Branch coloring in tree corresponds to genome name colors in key. Domain architectures typical of sequences in clustered branches ring the tree, see (A) for explanation of architecture depictions. Complete trees provided in Newick format in Supplemental Material.