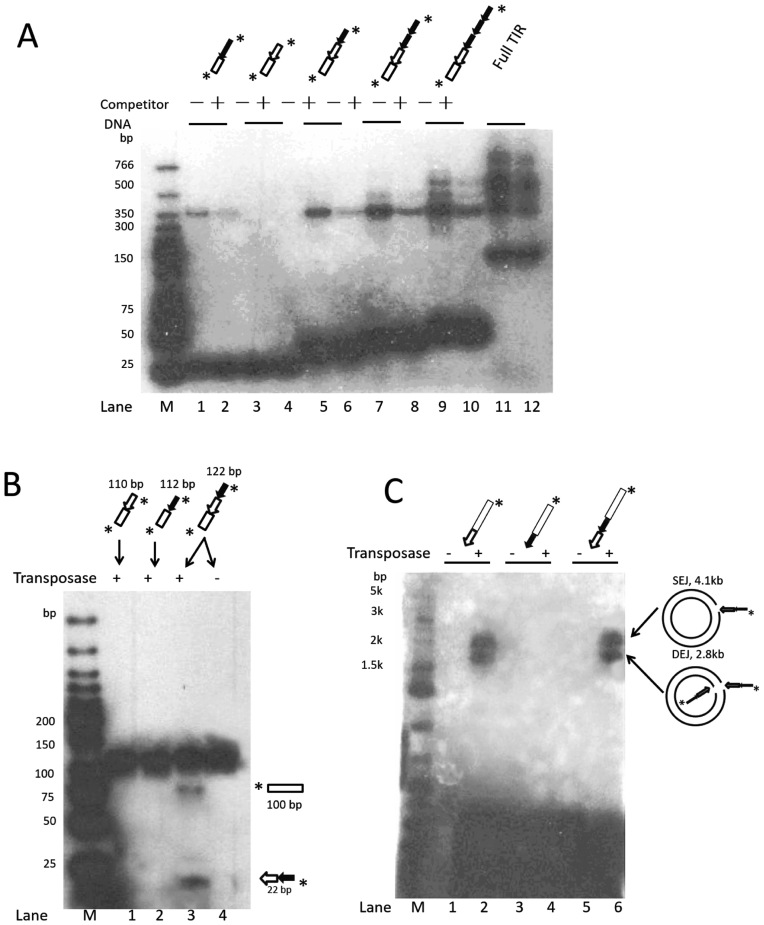
Figure 5.
In vitro analysis of the impact of the Muta1 subterminal repeats on transposase binding, double strand break and end joining reactions. (A) DNA binding assay. Diagrams at the top are structures of DNA substrates, all contain 20-bp flanking DNA segment (open box), subterminal repeats and terminal motif. The unlabeled 165-bp DNA containing 20-bp flanking DNA and the full length Muta1 L-TIR is added in 100 (+) molar excess as competitor DNA. Reaction products are resolved on a native polyacrylamide gel. Fast migrating bands are DNA substrates (30–165 bp). (B) Donor cleavage assay. Diagrams at the top are the structures of DNA substrates, diagrams at the bottom are predicted structures of cleavage products. Reaction products are resolved on a native polyacrylamide gel. (C) End joining assay. Diagrams at the top are the structures of DNA substrates, intact pUC19 plasmid is used as target DNA, reaction products are resolved on a native agarose gel.