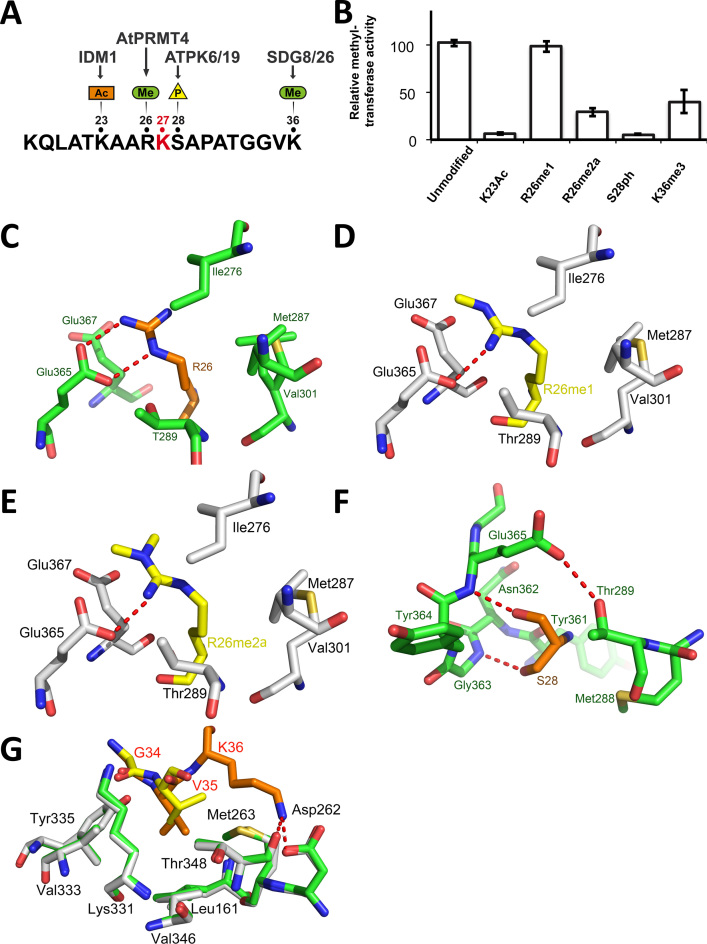
Figure 2.
Post-translational modifications of residues neighboring histone H3.1 K27 are detrimental to ATXR5 enzymatic activity. (A) Schematic representation of the H3.1 peptide showing the PTMs deposited in vicinity of the K27 methylation site (red). (B) Methyltransferase activity of ATXR5 SET domain on modified histone H3.1 peptides relative to un-modified H3.1. (C) Zoomed-view of ATXR5 SET domain binding cleft showing un-modified S28 (left) in which the carbon atoms of the peptides and proteins are colored in orange and green respectively. (D) Comparison of ATXR5/H3.1/AdoHcy with ATXR5/H3.1K36me3/AdoHcy in which the carbon atoms for the modified peptides and co-crystallized ATXR5 are colored in yellow and white, respectively. Zoomed-view of ATXR5 SET domain binding cleft showing un-modified R26 (E), R26me1 (F) and R26me2a (G). Carbon atoms of the un-modified and modified peptides are rendered in orange and yellow respectively. Hydrogen bonds are highlighted by red dashed lines.