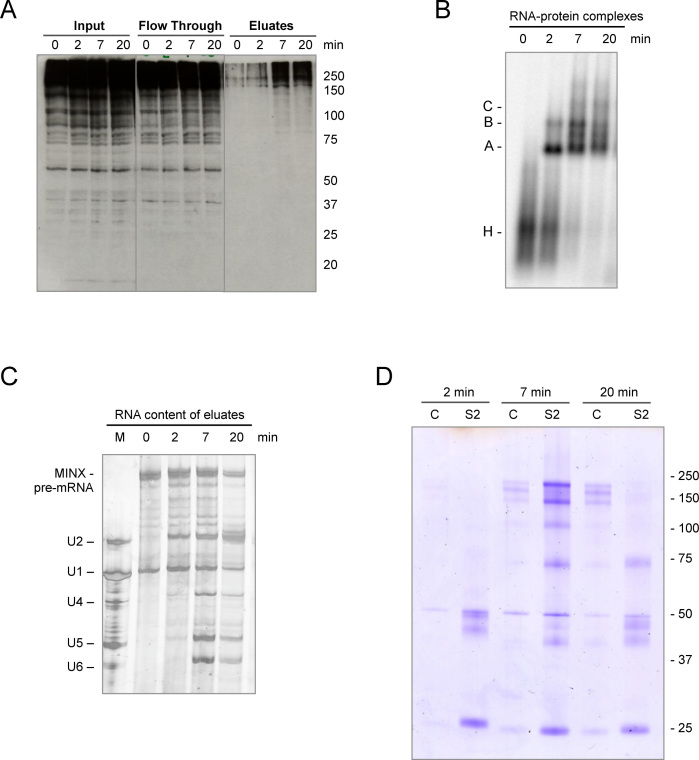
Figure 2.
Identification of SUMOylated proteins in spliceosomes formed in vitro. (A) Spliceosomal complexes were allowed to form on radiolabelled MINX pre-mRNA containing MS2 binding sites bound by MBP–MS2 fusion protein under in vitro splicing conditions for 0–20 min. The RNA–protein complexes were purified by MBP pull-down with amylose beads and, after elution with maltose, proteins in the purified spliceosomal complexes (eluates), as well as in the input and flow-through, were analyzed by WB with anti-SUMO2 antibody visualized by ECL reagent. (B) The kinetics of spliceosomal complex formation was monitored by native agarose gel electrophoresis. (C) RNA composition of affinity-purified splicing complexes was analyzed by denaturing PAGE. MINX-MS2 pre-mRNA and the U1-U6 snRNAs were visualized by silver staining. The marker (M) in lane 1: RNA from anti-m3G (2,2,7-trimethylguanosine-containing cap structure of snRNAs), affinity-purified human snRNPs. (D) SUMOylated proteins within the affinity-purified spliceosomal complexes were immunoprecipitated with an anti-SUMO2 antibody (S2), after disruption of the RNP complexes, and then analyzed by denaturing SDS-PAGE. As a control (C), immunoprecipitation was also performed with IgG.