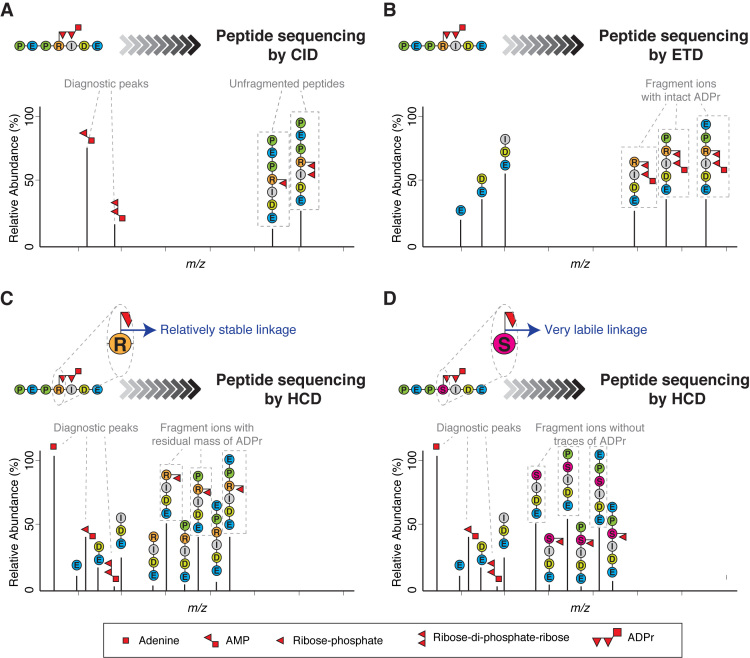
Figure 2.
Different fragmentation methods applied to ADP-ribosylated peptides. (A) Schematic representation of a typical collision-induced dissociation (CID) fragmentation spectrum from an ADP-ribosylated peptide. The CID spectrum contains more ions formed through fragmentation of the ADP-ribose itself than through fragmentation of the peptide backbone. As a consequence, it yields little or no information about the peptide sequence. However, CID can be used to confirm the presence of ADP-ribose on a precursor by generating modification-specific diagnostic peaks (e.g. AMP). (B) Schematic representation of a typical electron-transfer dissociation (ETD) fragmentation spectrum from an ADP-ribosylated peptide. The ETD spectrum is rich in information about the peptide sequence and the site of modification. A characteristic feature of ETD is the retention of the intact ADP-ribose. The masses of adjacent fragment ions differ by the masses of the corresponding amino acids. The presence of an ADP-ribose changes the mass of the modified amino acid residue and the ions containing the modified residue by 541.06 Da. (C) Schematic representation of a typical higher-energy collisional dissociation (HCD) fragmentation spectrum from an ADP-ribosylated peptide in which the ADP-ribose linkage to the modified amino acid is relatively stable under HCD fragmentation (e.g. ADPr on Arg). The HCD spectrum contains ions from fragmentation of both the peptide backbone and ADP-ribose. Usually, the HCD spectrum is dominated by one of the diagnostic ions generated by the internal fragmentation of ADP-ribose (adenine, at m/z 136.06), but additional fragments are also detected corresponding to the peptide with a residual part of ADP-ribose attached (typically ribose phosphate-H2O). These fragment ions provide the basis for determining the site of modification. (D) Schematic representation of a typical HCD spectrum from an ADP-ribosylated peptide in which the ADP-ribose linkage to the modified amino acid is very labile under HCD fragmentation (e.g. ADPr on Ser). The HCD spectrum contains ions from both the peptide backbone and from the ADP-ribose fragmentation. This spectrum can contain peptide fragment ions carrying part of ADP-ribose, but the strongest ion series is not that of a modified sequence, but of the native identified peptide. These de-modified fragment ions contain no information for determining the modification site. Since search algorithms score these de-modified ions as unmodified ions, conventional analysis of serine ADPr by HCD can lead to multiple, erroneous localizations arising from variations in spectrum quality. See Figure 3.