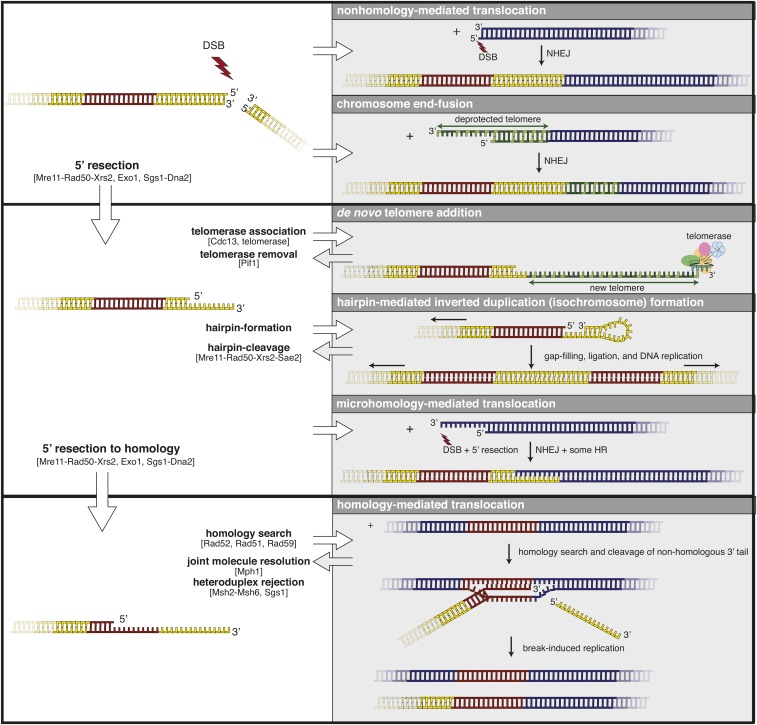
Figure 2.
DSBs can be acted on by a variety of DNA metabolic processes that can generate different kinds of GCRs. Top: DSBs lead to nonhomology-mediated translocations mediated by NHEJ and, in strains containing deprotected telomeres (green bases), can lead to dicentric chromosomes mediated by end-to-end fusions with other chromosomes (blue). Middle: 5′ resection of DSBs, which in mitotic cells is driven by a combination of Mre11-Rad50-Xrs2, Exo1, and Sgs1-Dna2, generates a 3′ overhang that is subject to several reactions. Cdc13 can recognize TG-rich ssDNA regions and promote the association of telomerase leading to synthesis of a de novo telomere; this association is antagonized by the activity of the Pif1 helicase. Overhang regions not bound by RPA can form hairpins. Upon gap filling by DNA polymerases, ligation, and a subsequent round of replication, these can give rise to dicentric (or centromere-less) inverted duplication chromosomes. These hairpins can also be cleaved through the action of Mre11-Rad50-Xrs2 in combination with Sae2. Resected ends can also lead to microhomology-mediated joining to other broken chromosomes resulting in translocations. Bottom: resection into sequences that have homology with other regions in the genome can lead to nonallelic HR, including BIR (data not shown) resulting in homology-mediated translocations. The Rad1-Rad10 nuclease has been implicated in the removal of the nonhomologous 3′-tail (yellow) during these types of events. BIR, break-induced replication; DSBs, double-strand breaks; GCRs, Gross Chromosomal Rearrangements; HR, homologous recombination; NHEJ, nonhomologous end joining; RPA, Replication Protein A.