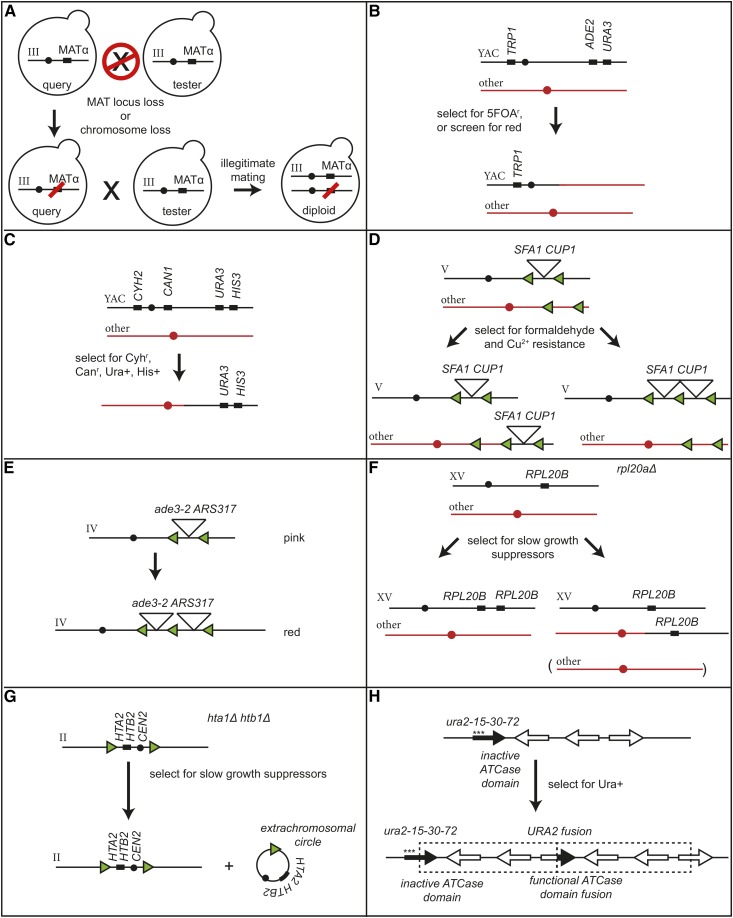
Figure 6.
Comparison of haploid GCR assays that select for amplification of markers or loss of internal markers. (A) The “a-like faker” mating assay that uses mating and selection to detect loss of information at the MAT locus (Lemoine et al. 2005; Yuen et al. 2007). (B) A YAC-based GCR assay that selects for loss of URA3 and ADE2 but retention of TRP1, which also detects translocations involving fragments of native S. cerevisiae chromosomes (Huang and Koshland 2003; Wahba et al. 2013). (C) A YAC-based GCR assay that selects for retention of the terminal YAC arm and loss of the remainder of the chromosome, and detects translocations involving fragments of native S. cerevisiae chromosomes (Tennyson et al. 2002). (D) Amplification of a SFA1-CUP1 cassette causes increased resistance to formaldehyde and copper ions that detects amplification resulting from HR-mediated unequal crossing over (Zhang et al. 2013a). (E) Amplification of ade3-2 resulting from HR-mediated unequal crossing over causes S. cerevisiae colonies to undergo a color change from pink to red (Green et al. 2010). (F) An assay for suppressors of the slow growth phenotype of an rpl20aΔ mutant strain selects for amplification of RPL20B (Koszul et al. 2004; Payen et al. 2008). (G) An assay for suppressors of the slow growth phenotype of an hta1Δ htb1Δ double-mutant strain selects for amplification of the centromere (black circle) proximal HTA2 and HTB2 resulting from the formation of a circular chromosome (Libuda and Winston 2006). (H) Reactivation of ura2-15-30-72, which has three nonsense mutations in the 5′-end of the gene (asterisks), by selecting for uracil prototrophy has identified the formation of large duplications in which the 3′-end of the ura2 allele is fused in-frame to another open reading frame resulting in expression of a functional Ura2 fusion protein (Schacherer et al. 2005). ATCase, aspartate carbamyltransferase; CanR, canavanine-resistant; GCRs, Gross Chromosomal Rearrangements.