Abstract
The essential histone chaperone FACT plays a critical role in DNA replication, repair, and transcription, primarily by binding to histone H2A-H2B dimers and regulating their assembly into nucleosomes. While FACT histone chaperone activity has been extensively studied, the exact nature of the H2A and H2B residues important for FACT binding remains controversial. In this study, we characterized the functions of residues in the histone H2A and H2B acidic patch, which is important for binding many chromatin-associated factors. We found that mutations in essential acidic patch residues cause a defect in histone occupancy in yeast, even though most of these histone mutants are expressed normally in yeast and form stable nucleosomes in vitro. Instead, we show that two acidic patch residues, H2B L109 and H2A E57, are important for histone binding to FACT in vivo. We systematically screened mutants in other H2A and H2B residues previously suspected to be important for FACT binding and confirmed the importance of H2B M62 using an in-vivo FACT-binding assay. Furthermore, we show that, like deletion mutants in FACT subunits, an H2A E57 and H2B M62 double mutant is lethal in yeast. In summary, we show that residues in the nucleosome acidic patch promote histone occupancy and are important for FACT binding to H2A-H2B dimers in yeast.
Keywords: nucleosome assembly, Spt16, Pob3, Nap1, H2A-H2B dimer
EUKARYOTIC DNA is assembled into nucleosomes by wrapping ∼147 bp of DNA around an octamer of histone proteins containing two H2A-H2B dimers and an H3-H4 tetramer (Luger et al. 1997). Within the cell, nucleosome assembly (and disassembly) is greatly facilitated by the action of histone chaperone proteins (Das et al. 2010; Gurard-Levin et al. 2014). Histone chaperones bind free histones to prevent the formation of nonspecific histone-DNA aggregates, and often deliver their bound histones to sites of active nucleosome assembly, such as at replication forks (Andrews et al. 2010; Burgess and Zhang 2013; Gurard-Levin et al. 2014). Histone chaperones can also facilitate the nucleosome disassembly and reassembly associated with RNA polymerase transcription and DNA repair (Avvakumov et al. 2011); however, for many histone chaperones, the molecular details of histone binding and chaperone activity are not fully understood.
FACT (facilitates chromatin transcription) is an essential histone H2A-H2B chaperone that is primarily implicated in nucleosome disassembly and reassembly during transcription, but also regulates nucleosome dynamics during DNA replication and the DNA damage response (Formosa 2012; Polo 2015). FACT is comprised of two essential subunits: Spt16 and Pob3 (SSRP1 in mammalian cells), which are present in essentially all eukaryotes. FACT associates with nucleosomes in part through its DNA-binding HMG-box domain, located in the SSRP1 subunit in mammalian cells and in the nonessential accessory Nhp6 subunit in yeast (Allain et al. 1999; Orphanides et al. 1999; Formosa et al. 2001; Masse et al. 2002; Winkler et al. 2011). FACT also binds histone proteins, particularly histones H2A and H2B, although the exact histone regions required for FACT binding are controversial (Hondele et al. 2013; Kemble et al. 2015). A previous structural study suggested that a region in the H2B α1 helix is important for binding to the Spt16 middle domain (Hondele et al. 2013). In contrast, a more recent study indicated that the Spt16 middle domain is not important for H2A and H2B binding, but instead the Spt16 and Pob3 acidic tails bind a small hydrophobic pocket associated with the H2B α2 helix (Kemble et al. 2015). However, both of these binding targets are normally inaccessible for FACT binding in chromatin, as they are located at the histone–DNA binding interface of nucleosomes and are thus sterically occluded by the nucleosomal DNA.
While histone proteins have an overall positive charge to facilitate efficient binding to DNA, there is a region on the surface of the nucleosome deemed the “acidic patch” that is formed by multiple aspartic and glutamic acid residues in histone H2A and H2B (H2A E57, E62, D65, D91, E92, E93, and H2B E116), as well as other, nonacidic residues, including H2A Y58, H2A L66, and H2B L109. Notably, all the H2A and H2B residues essential for yeast survival (H2A Y58, E62, D91, and H2B L109) are located in the acidic patch, but their essential functions are unclear. The acidic patch serves as an important interface for internucleosome interactions, as it interacts with the H4 tail of the neighboring nucleosome (Luger et al. 1997; Kalashnikova et al. 2013; Wilkins et al. 2014). Additionally, recent evidence shows the acidic patch is important for interactions with nonhistone proteins (Wyrick et al. 2012), including regulator of chromosome condensation 1 (RCC1) (Makde et al. 2010), the yeast silencing protein (Armache et al. 2011), IL-33 (Roussel et al. 2008), HMGN2 (Kato et al. 2011), polycomb repressive complex 1 (PRC1) (McGinty et al. 2014), the SAGA histone deubiquitylating complex (Morgan et al. 2016), and the Set8 histone methyltransferase (Girish et al. 2016), as well as other proteins. However, it is unclear if the essential function of the acidic patch in yeast entails mediating interactions with these and other chromatin proteins.
In the course of characterizing the in-vivo functions of essential acidic patch residues in yeast, we discovered that essential acidic patch residues H2A D91 and H2B L109 are important for maintaining wild-type (WT) levels of histone occupancy. Importantly, this decrease in histone occupancy was not due to an intrinsic defect in nucleosome stability. Instead, co-immunoprecipitation (co-IP) assays indicate that acidic patch residues H2B L109 and H2A E57 are important for FACT binding to free H2A-H2B dimers in yeast, a conclusion that is corroborated by genetic analysis of these and other histone mutants.
Materials and Methods
Yeast strains and plasmids
Histone mutant plasmids were constructed from pJW500 (Hodges et al. 2015) using a modified version of the QuickChange protocol, and mutations were verified by DNA sequencing. Yeast strains containing viable histone mutants as the only histone copy were grown in YPD media; strains containing two histone plasmids were grown in double-selective SC media to maintain both plasmids. Isogenic WT strains were used as controls for all experiments. Yeast strain information can be found in Supplemental Material, Table S1 in File S1.
Yeast spotting
Indicated plates were spotted with 10-fold serial dilutions of yeast. The FLO8-HIS3 reporter strain was used as in Hodges et al. (2015). Pictures were taken after incubation at 30° for 4 days (5-FOA; HU) or 7 days (FLO8-HIS3).
Yeast growth curve analysis
Growth curve analysis was performed as in Jin et al. (2007) and Hodges et al. (2015). Briefly, plasmids containing WT or mutant histones were transformed into yeast containing WT histones under control of the Gal promoter and grown in media containing galactose. Cells were then switched to glucose media to shut down the WT histones. OD600 measurements were taken at time points following the switch to glucose (time 0).
Chromatin IP
Chromatin IP (ChIP) experiments were conducted as described previously (Hodges et al. 2015). Briefly, log-phase yeast culture was cross-linked using 1% formaldehyde at room temperature for 15 min followed by quenching with 0.125 M glycine. After cell lysis by bead beating, chromatin was sonicated using the Bioruptor (Diagenode) to fragments averaging ∼400–500 bp. Chromatin was immunoprecipitated using an anti-HA (Thermo) or anti-FLAG (Sigma Chemical, St. Louis, MO) antibody conjugated to magnetic beads (Invitrogen, Carlsbad, CA). After elution, samples were proteinase-K digested and cross-links were reversed at 65° overnight. DNA was purified with phenol and phenol/chloroform/isoamyl alcohol and RNAse A digested before analysis by quantitative PCR (qPCR) using the ABI Fast 7500 qPCR system and EvaGreen reagents (Biotium). Detailed qPCR information, including primer sequences, can be found in the minimum information for publication of quantitative PCR experiments (MIQE) in Table S2 in File S1.
Co-IP
Co-IP analysis was conducted as in Mao et al. (2016) with some modifications, using Spt16-9xMyc or Nap1-9xMyc yeast strains. Briefly, 50 ml of yeast culture was collected, then resuspended in ice-cold co-IP lysis buffer [40 mM HEPES, pH 7.5, 0.1% Tween 20, 150–200 mM NaCl, 10% glycerol, 1× Protease Inhibitor Cocktail (Roche)]. Cells were lysed by bead beating, then, if applicable, whole-cell extracts were treated with 0.32 U/μl Pierce Universal Nuclease (Catalog number PI88700) for 30 min at 4°. The extract was centrifuged, and the soluble fraction was used for co-IP with anti-Myc beads (Thermo) overnight at 4°. After washing, proteins were eluted from the beads by boiling in 1× SDS-PAGE buffer. IP and input samples were analyzed by Western blot using antibodies against H2A (Active Motif), H2B (Active Motif), HA (Thermo), and c-Myc (9E10; Santa Cruz Biotechnologies). Blots were scanned using the Typhoon FLA 7000 (GE Healthcare) and analyzed using ImageQuant software. Graphs represent the mean and SD of at least three independent replicates; P-values were calculated using an unpaired t-test.
Data availability
Yeast strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Phenotypic characterization of essential acidic patch residues in yeast
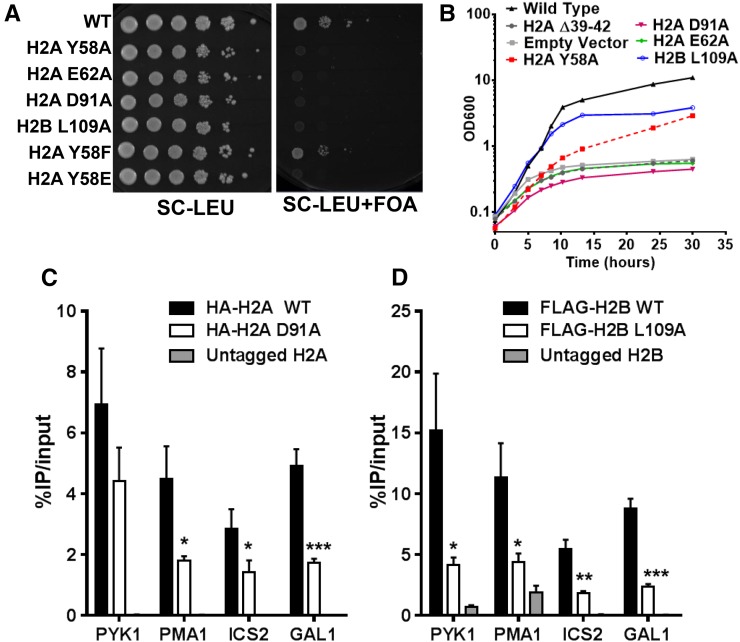
It has been previously reported that residues H2A Y58, E62, D91, and H2B L109 in the acidic patch are essential for yeast survival, as alanine mutants in any of these residues were lethal in yeast (Matsubara et al. 2007; Nakanishi et al. 2008; Sakamoto et al. 2009), which we confirmed in our strain background (S288C; Figure 1A). To investigate the essential functions of these acidic patch residues, we first analyzed the kinetics of growth arrest in yeast strains expressing acidic patch mutants using a galactose (Gal) shutdown system (Jin et al. 2007; Hodges et al. 2015). In this experimental system, both WT and mutant histones are expressed on separate plasmids: one plasmid contains WT histones, with the histone H2A and H2B genes under the control of the GAL1-10 promoter; and the second plasmid contains either WT or mutant histones under control of their native promoters. In media containing galactose, histones from both plasmids are expressed; in glucose media, WT H2A and H2B expression is rapidly repressed, enabling us to characterize the kinetics of growth arrest for the lethal acidic patch mutants.
Figure 1.
(A) Phenotypes of mutants in essential acidic patch residues. Yeast cells containing the indicated WT or mutant histone genes present on a LEU2 plasmid were spotted on SC-LEU or SC-LEU+FOA media. A URA3 plasmid containing WT histone genes was shuffled out by 5-FOA selection. (B) Growth curve analysis in a Gal shutdown system. WT histone expression was repressed upon shift to glucose media (see Materials and Methods) at time 0, allowing only the indicated histone mutants to be expressed. A representative growth curve is shown. (C and D) ChIP-qPCR analysis of histone occupancy in a two-plasmid system. HA-H2A or FLAG-H2B WT (black) or mutant (white) histones were coexpressed with WT untagged histones and immunoprecipitated using the corresponding anti-HA or anti-FLAG antibody. Untagged WT histones (gray) were used as a negative control. Data are shown as percent IP normalized to input whole-cell extracts. P-values were calculated for the represented triplicate experiment using an unpaired t-test. Mean ± SD is depicted. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, and **** P ≤ 0.0001.
We analyzed the kinetics of growth arrest of yeast cells containing an empty vector (i.e., no H2A or H2B expression) or an H2A Loop1 deletion (H2A ∆39–42), which is expected to disrupt H2A folding and nucleosome assembly (Hodges et al. 2015). Both control strains had a rapid growth arrest after the shift to glucose (Figure 1B), consistent with our previously published data (Hodges et al. 2015). In contrast, a strain expressing WT H2A and H2B grew normally after the shift to glucose. Cells expressing mutants of essential acidic residues within the acidic patch (H2A E62A and D91A) had a rapid growth arrest following the shift to glucose, with arrest kinetics that mirrored those of the empty vector and H2A Loop1 deletion controls (Figure 1B). However, cells expressing mutants in essential uncharged residues (H2A Y58A and H2B L109A) had a delayed growth arrest following the switch to glucose (Figure 1B), continuing to grow for >10 hr. The essential acidic patch residues stratified into two distinct phenotypic classes: mutants in charged acidic patch residues (i.e., H2A E62 and D91) had a rapid and severe growth arrest, while mutants in uncharged acidic patch residues (i.e., H2A Y58 and H2B L109) had a delayed growth arrest.
Mutations in essential acidic patch residues cause defects in histone occupancy in yeast, but do not affect intrinsic nucleosome stability in vitro
The rapid and severe growth arrest of mutants in the charged acidic patch residues suggests that these acidic patch mutants affect histone expression, folding, or nucleosome assembly. To test this hypothesis, we first examined whether mutants in essential acidic patch residues altered histone expression. The acidic patch mutants were constructed in HA-tagged histone H2A or FLAG-tagged histone H2B, and coexpressed in yeast with untagged WT H2A-H2B to maintain cell viability. The presence of the tag on the mutant histone allowed for separation of the mutant and WT histones during PAGE and both were detected using histone-specific antibodies (Figure S1 in File S1).
Western blot analysis indicated that the H2A E62A and H2B L109A mutants were expressed at similar levels as WT H2A and H2B (Figure S1 in File S1). The expression levels of the H2A Y58A and D91A mutant histones appeared to be significantly reduced relative to WT using an anti-H2A antibody (Figure S1A in File S1, top panel); however, when we used HA-specific antibodies to measure the expression, the HA-tagged H2A D91A mutant was expressed at normal levels (Figure S1A in File S1, bottom panel). Based on these data, it appears that the D91A mutant does not affect H2A protein expression, but rather antibody recognition. This is in accordance with a previous study, which found that mutants in neighboring H2A residues (i.e., H2A E93A and H2A L94A) also had lower H2A signal by Western blot using the same anti-H2A antibody (Cucinotta et al. 2015). In contrast, H2A Y58A could not be detected by Western blot using anti-H2A or anti-HA antibodies (Figure S1A in File S1), suggesting this mutant is not stably expressed in yeast. Since H2A Y58 is phosphorylated in yeast and mammalian cells (Basnet et al. 2014), we also measured the expression of H2A Y58 mutants that would eliminate or mimic Y58 phosphorylation (H2A Y58F and Y58E, respectively). Western blotting using anti-H2A and anti-HA antibodies revealed that the H2A Y58F mutant is expressed normally in yeast (Figure S1A in File S1), while the H2A Y58E mutant protein level is decreased relative to WT H2A (Figure S1A in File S1), which may explain why the H2A Y58E mutant is lethal in yeast (Figure 1A).
We next examined histone occupancy in yeast using a two-plasmid system (Hodges et al. 2015; Mao et al. 2016) to measure histone occupancy of lethal acidic patch mutants. We selected one essential residue from each class, charged (H2A D91) and uncharged (H2B L109), for our analysis. An HA-tagged H2A D91A mutant was coexpressed with untagged WT H2A to maintain strain viability. ChIP analysis using an anti-HA antibody was used to specifically measure occupancy of the HA-H2A D91A mutant. Similar experiments were performed for the H2B L109A mutant, except H2B was FLAG-tagged.
We analyzed H2A or H2B occupancy by ChIP-qPCR using anti-HA or anti-FLAG antibody at four representative yeast loci: the 5′ coding regions of GAL1 and PYK1, the coding region of PMA1, and promoter of ICS2 (Hodges et al. 2015). HA-H2A occupancy for H2A D91A was significantly enriched relative to the untagged control at all four loci tested (Figure 1C), indicating that the H2A D91A mutant histone can be assembled into chromatin in vivo. However, the level of HA-H2A occupancy for the H2A D91A mutant was generally lower than the WT control (HA-H2A WT, Figure 1C). ChIP analysis of FLAG-H2B L109A occupancy yielded similar results. FLAG-H2B occupancy for H2B L109A was enriched relative to the untagged control, but the level of FLAG-H2B L109A occupancy was approximately three- to fourfold lower than WT at all four loci tested (Figure 1D). In general, the H2B L109A mutant caused a somewhat greater decrease in histone occupancy than the H2A D91A mutant, particularly at the PYK1 locus. These results indicate that essential acidic patch residues in both classes (charged and uncharged side chains) are important for normal levels of histone occupancy in yeast.
Although these acidic patch residues are located on the solvent-exposed nucleosome surface, it is formally possible that the decrease in histone occupancy in vivo could reflect an intrinsic instability of nucleosomes containing mutants in essential acidic patch residues. Previous studies have shown that a quadruple acidic patch mutant (H2A E61A, E64A, D90A, and E92A in Xenopus; corresponding to H2A E62A, E65A, D91A, and E93A in yeast) formed stable nucleosomes in vitro (Kim et al. 2015), indicating that essential, charged acidic patch residues do not significantly affect nucleosome stability. However, the effect of H2B L109 mutants on nucleosome stability has not been previously examined.
To characterize the effect of the H2B L109A mutant on nucleosome stability, we prepared nucleosomes from recombinant Xenopus histones containing the H2B L103A mutant (homologous to H2B L109A mutant in yeast). Nucleosome core particle (NCP) stability was compared by salt-induced dissociation monitored by a previously described protein–protein fuorescence resonance energy transfer (FRET) system (Hoch et al. 2007). This FRET system was designed to specifically report on the dissociation of the H2A-H2B dimers from the NCP; the FRET signal is largely insensitive to DNA breathing and does not report on interactions between the (H3-H4)2 tetramer and DNA. The L103A mutation had a minor effect on the apparent cooperativity of dissociation of the heterodimers, suggesting that the hexasome (in which only one H2A-H2B dimer has dissociated) may be slightly destabilized relative to the WT NCP (Figure S2 in File S1). However, the initial portion of the transition, reflecting dissociation of the first H2A-H2B dimer, was not affected by the mutation (Figure S2 in File S1). In summary, these data indicate the H2B-L103A mutation has only minor effects on NCP stability in vitro, which cannot explain the defect in histone occupancy observed in vivo.
Acidic patch residues H2B L109 and H2A E57 are important for histone H2A and H2B binding to FACT
Nucleosome assembly in vivo typically requires the activity of histone chaperone proteins. Since mutations of the essential acidic patch residues caused a defect in nucleosome occupancy, we tested whether these residues were important for binding to the H2A-H2B chaperones FACT and Nap1. We performed co-IP analysis using myc-tagged Nap1 or Spt16 (a subunit of FACT) from yeast whole-cell extract, following our published protocol (Mao et al. 2016). Since relatively little DNA is present in the soluble whole-cell extract fraction used in these experiments (Mao et al. 2016), the co-IP results presumably reflect Spt16 or Nap1 binding to soluble, free histones (e.g., H2A-H2B dimers). To analyze the binding of mutant acidic patch residues, we coexpressed a FLAG- or HA-tagged mutant histone with WT histones. The FLAG or HA tag causes a band shift detectable by Western blot using a histone-specific antibody to distinguish the mutant histone from untagged WT histone, which was coexpressed to maintain cell viability. The presence of WT histone also serves as an internal control, allowing for a direct comparison of binding between WT and mutant histones.
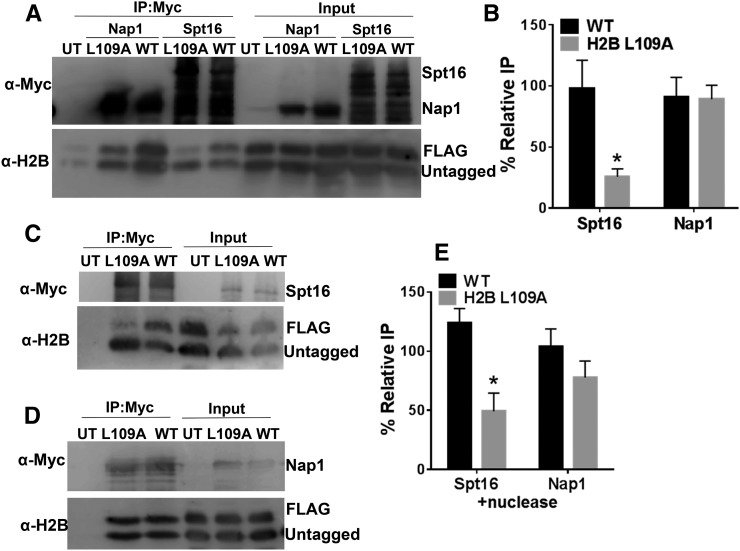
The co-IP data indicated that both Nap1 and Spt16 bound histone H2B (Figure 2), in accordance with previous results (e.g., Foltman et al. 2013; Mao et al. 2016); however, the H2B L109A mutant significantly decreased H2B binding to Spt16 (Figure 2, A and B). In the Spt16 IP sample, the band corresponding to the FLAG-H2B L109A mutant had significantly lower intensity than the untagged WT H2B band (compare top and bottom bands in αH2B blot in lane 4 of Figure 2A). In contrast, FLAG-H2B WT was co-immunoprecipitated with Spt16 at a similar level as untagged H2B WT (compare top and bottom bands in lane 5 of Figure 2A). Quantification of replicate Spt16 IP samples indicated that, on average, FLAG-H2B L109A bound Spt16 approximately fourfold less than untagged WT H2B (Figure 2B; P < 0.05), and no difference in binding was observed between FLAG-H2B WT and untagged WT H2B. While H2B L109A significantly decreased H2B binding to Spt16, there was no significant difference in the amount of the FLAG-H2B L109A bound to Nap1 (compare top and bottom bands in lane 2 of Figure 2A). These data indicate that H2B L109 residue is important for histone H2B binding to FACT but not Nap1 in vivo.
Figure 2.
H2B L109A mutant causes a defect in H2B binding to FACT. (A) Co-IP analysis using anti-myc beads of yeast whole-cell extracts containing myc-tagged Spt16 or Nap1. FLAG-tagged mutant histones were coexpressed with untagged WT histones and separated by Western blot. The anti-myc Western blot detects multiple bands (likely representing Spt16-myc degradation products), as observed previously (Mao et al. 2016), because the lane is overloaded to detect the relatively low levels of soluble histones bound to Spt16. (B) Quantification of the percent IP of the tagged band relative to the untagged band. Graphs represent the mean and SEM of at least three independent replicates. (C–E) Nuclease treatment does not improve H2B L109A binding to (C) Spt16 or (D) Nap1. Cell extracts were treated with 0.32 U/μl Pierce universal nuclease prior to IP to release chromatin-bound histones and increase the soluble histone pool. (E) Quantification of the percent IP of the tagged band relative to the untagged band. Graphs represent the mean and SEM of at least three independent replicates. * P < 0.05.
This analysis was somewhat complicated by the relatively low levels of histone binding to FACT in vivo. However, previous reports have shown that FACT binds strongly to histones released from chromatin by nuclease treatment prior to the co-IP step (Foltman et al. 2013; Mao et al. 2016). Hence, we examined whether the H2B L109A mutant affected FACT binding under these conditions. As expected, we observed elevated histone binding to Spt16, but not Nap1, following nuclease treatment (data not shown). However, while overall H2B binding to Spt16 increased following nuclease treatment, the binding of the H2B L109A mutant to Spt16 remained significantly lower than WT under these conditions (compare top and bottom bands in αH2B blot in lane 2 of Figure 2C). Analysis of replicate Spt16 IP samples indicated that binding of the H2B L109A mutant to Spt16 was about threefold lower than WT, even following nuclease treatment (Figure 2E; P < 0.05). Again, there was no significant difference in H2B L109A binding to Nap1 following nuclease treatment (Figure 2, D and E). These data confirm that H2B L109 residue is important for histone binding to FACT, but not to Nap1.
While H2B L109 is important for FACT binding, our data indicate that mutants in other essential acidic patch residues did not significantly affect FACT binding to histone H2A and H2B. We observed no significant difference in the amount of Spt16-bound HA-H2A E62A or HA-H2A Y58F relative to the untagged H2A control or HA-H2A WT (Figure S3 in File S1). Since H2A D91A affects recognition by the anti-H2A antibody, we analyzed H2A D91A using the HA antibody, which showed H2A D91A also binds FACT at WT levels (Figure S3 in File S1).
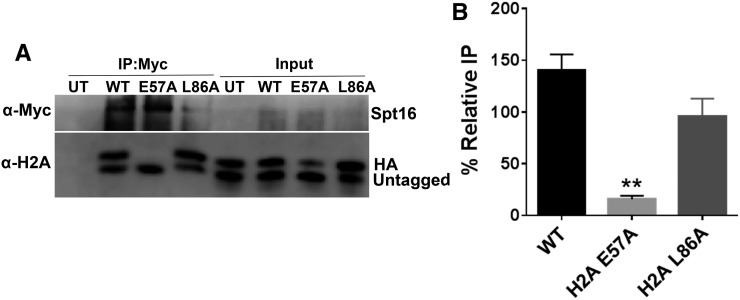
A recent study (Cucinotta et al. 2015) reported that alanine mutants in nonessential acidic patch residues H2A E57 and H2A E93 decrease the recruitment of FACT to a number of actively transcribed genes in yeast. However, it is not known if decreased FACT recruitment is caused by the impaired FACT binding to these histone mutants. We measured binding of these mutant histones to Spt16 using our co-IP assay. As a control, we also tested H2A L86A, a nearby nonessential residue that, when mutated, enhances FACT recruitment to chromatin (Cucinotta et al. 2015). To directly compare effects of essential and nonessential acidic patch residues, and to mitigate the slow-growth phenotype of the H2A E57A and L86A mutants (data not shown), we HA-tagged the mutant histone and coexpressed untagged WT H2A, as described above. Since H2A signal was relatively weak following co-IP of Spt16, we treated yeast whole-cell extracts with nuclease to release additional histones from the chromatin into the soluble fraction (see above).
The co-IP data indicated that the histone H2A E57A mutant severely decreased FACT binding to histone H2A (Figure 3). While WT H2A was efficiently immunoprecipitated with Spt16, HA-H2A E57A was nearly absent in the Spt16 IP samples (compare HA-H2A bands in lanes 2 and 3 in Figure 3A). Quantification of replicate Spt16 IP samples confirmed that the H2A E57A mutant is not efficiently bound by Spt16 in the presence of untagged WT, even after normalizing for the somewhat lower levels of HA-H2A E57A in the input (Figure 3B; P < 0.01). In contrast, the H2A L86A mutant had only a marginal effect, if any, on Spt16 binding in this assay (Figure 3). Similar experiments were performed on the H2A E93A mutant, but we did not obtain consistent Spt16 co-IP data for this mutant (data not shown). In summary, these experiments indicate that two acidic patch residues, H2B L109 and H2A E57, are important for FACT binding to the H2A-H2B dimer.
Figure 3.
The acidic patch residue H2A E57 is important for binding FACT. (A) Co-IP analysis using anti-myc beads of nuclease-treated whole-cell extracts containing Spt16-myc and HA-tagged WT or indicated mutant histones. Untagged WT histones are coexpressed. (B) Quantification of the percent IP of the tagged band relative to the untagged band. Graphs represent the mean and SEM of at least three independent replicates. ** P < 0.01.
Previously identified H2A-H2B target residues also affect FACT binding in vivo
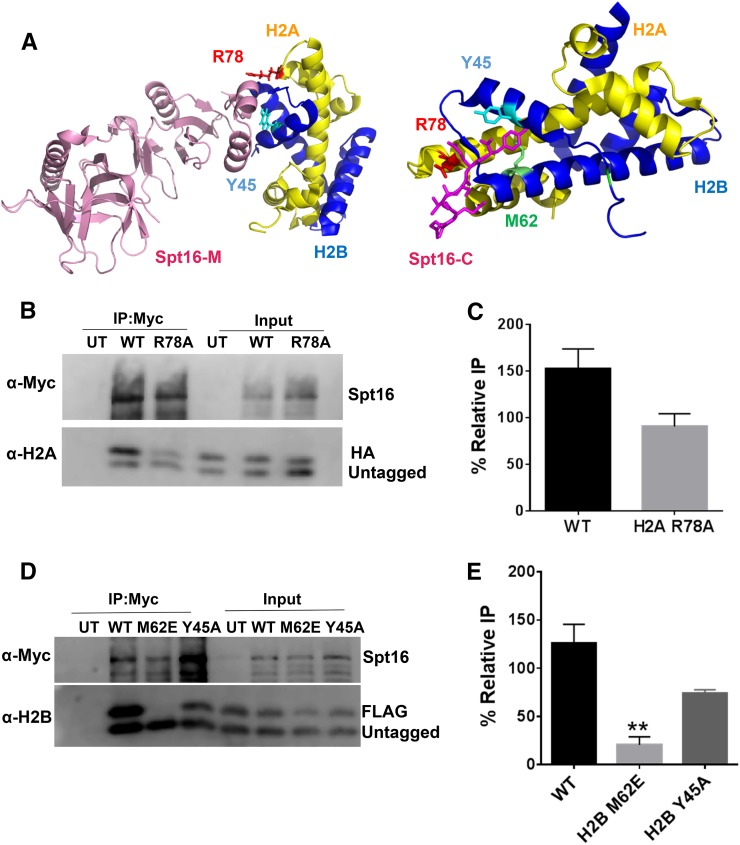
We next used our co-IP assay to compare the effects on FACT binding of other established or hypothesized FACT-binding targets in H2A and H2B, based on recently published FACT-histone crystal structures and in-vitro biochemical data (Hondele et al. 2013; Kemble et al. 2015). These residues in yeast are H2A R78, H2B Y45 (identified in both studies), and H2B M62 (Kemble et al. 2015) (see Figure 4A). One additional residue in Xenopus H2B (H2B I36) was also shown to be important for Spt16 binding (Hondele et al. 2013), but this residue is not conserved in yeast (corresponds to yeast H2B S42). While H2A R78, H2B Y45, and H2B M62 were shown in these studies to be important for Spt16 binding in vitro (Hondele et al. 2013; Kemble et al. 2015), it is not known to what extent they affect binding to FACT in vivo.
Figure 4.
Analysis of FACT binding to histone mutants identified from structural studies. (A) Published crystal structures of FACT in complex with histones pdbID#4KHA (left) and pdbID#4WNN (right). Residues identified as important for FACT binding (e.g., H2A R78, H2B Y45, and H2B M62) are indicated. Image was created using PyMOL. (B and C) H2A R78A does not inhibit binding to FACT. (B) Co-IP analysis using anti-myc beads of nuclease-treated Spt16-myc yeast whole-cell extracts containing HA-tagged WT or H2A R78A mutant histones. An untagged WT is coexpressed. (C) Quantification of the percent IP of the tagged band relative to the untagged band. Graphs represent the mean and SEM of at least three independent replicates. (D and E) H2B M62E significantly inhibits FACT binding. (D) Co-IP analysis using anti-myc beads of yeast whole-cell extracts containing myc-tagged Spt16 and FLAG-tagged WT H2B, H2B M62E, or H2B Y45A mutant histones coexpressed with untagged WT histones. (E) Quantification of the percent IP of the tagged band relative to the untagged band. Graphs represent the mean and SEM of at least three independent replicates. ** P < 0.01.
We tested the effects of H2A R78A, H2B Y45A, and H2B M62E mutants on FACT binding in yeast using our co-IP assay. The experimental design was similar as for the acidic patch residues, in that tagged H2A or H2B containing the indicated mutant was coexpressed with untagged WT H2A and H2B in yeast, and myc-tagged Spt16 was immunoprecipitated following nuclease treatment of yeast chromatin. The co-IP data indicated that the H2A R78A mutant had only a marginal effect, if any, on H2A binding to FACT (Figure 4, B and C, and Figure S4 in File S1). A previous study indicated that the H2A R78E mutant had a more severe Spt16 binding defect than the H2A R78A mutant in vitro (Kemble et al. 2015). We constructed a yeast strain containing the H2A R78E mutant, but found that it is lethal in yeast (Figure S5A in File S1) and does not express well by Western blot even when coexpressed with WT H2A (Figure S5B in File S1), suggesting this mutation affects histone expression or stability in yeast. While the H2A R78A mutant had only a marginal effect on binding to FACT, our co-IP data indicate that this mutant significantly decreased H2A binding to Nap1 (Figure S4 in File S1). Hence, our data suggest that H2A R78 is largely dispensable for binding to FACT, but is required for binding to Nap1.
The H2B M62E mutant significantly decreased H2B binding to FACT (Figure 4, D and E). There was significantly less FLAG-H2B M62E mutant immunoprecipitated with Spt16 than either untagged WT H2B or FLAG-H2B WT (Figure 4D). Quantification of replicate experiments revealed that H2B binding to Spt16 decreased at least fourfold in the H2B M62E mutant relative to WT (Figure 4E; P < 0.01). We also observed a marginal decrease in H2B binding to Spt16 in the H2B Y45A mutant (Figure 4, D and E). The defect in H2B binding to Spt16 was much greater in magnitude with H2B M62E relative to H2B Y45A, which is consistent with results from in-vitro binding studies (Kemble et al. 2015). Neither H2B Y45A nor H2B M62E significantly affected H2B binding to Nap1 when these mutants were expressed as the sole H2B copy in yeast (Figure S6, A and B, in File S1). A comparison of the Spt16 binding data indicated that mutants in acidic patch residues H2B L109A and E57A caused a roughly similar defect in H2A-H2B binding to FACT as H2B M62E and Y45A (previously identified binding targets), with H2A E57A and H2B M62E having the greatest effect on FACT binding.
Histone residues important for FACT binding have phenotypes similar to FACT mutants
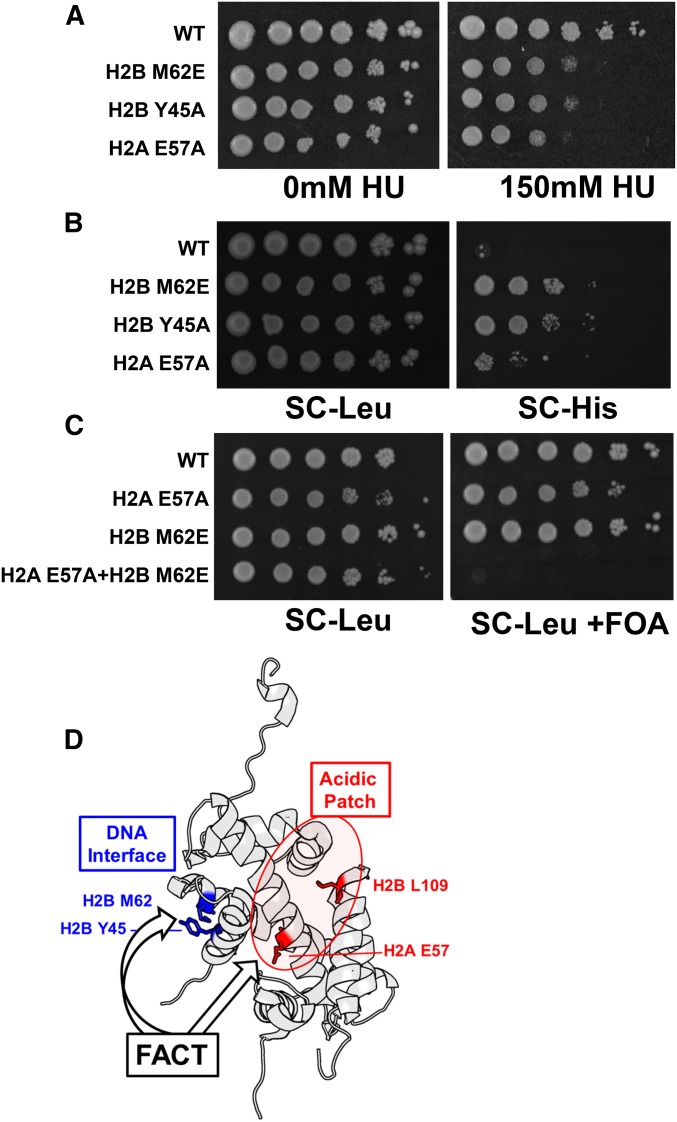
While Spt16 and Pob3 are essential for yeast viability, partial loss-of-function mutants in these FACT subunits have been previously shown to be sensitive to the replication inhibitor hydroxyurea (HU) and to induce cryptic transcription in FLO8 and other yeast genes (Kaplan et al. 2003; Cheung et al. 2008; Formosa 2012). We tested whether histone residues important for FACT binding in vivo had similar mutant phenotypes in yeast. Our data indicate that the H2B Y45A, M62E, and H2A E57A single mutants were sensitive to HU (Figure 5A), consistent with previous reports (Matsubara et al. 2007; Kemble et al. 2015). To characterize the effects of these mutants on FLO8 cryptic transcription, we used a FLO8::HIS3 reporter gene, in which the HIS3 ORF was inserted downstream of the FLO8 cryptic internal promoter (Cheung et al. 2008; Hodges et al. 2015). The WT strain was unable to grow on media lacking histidine (SC-His; Figure 5B), indicating that cryptic transcription of FLO8::HIS3 reporter was repressed. In contrast, H2B Y45A, M62E, or H2A E57A mutants showed enhanced growth on SC-His media (Figure 5B), indicating that these mutants induced FLO8 cryptic transcription.
Figure 5.
Histone H2A and H2B residues important for FACT binding have yeast phenotypes in common with FACT mutants. (A) Serial dilutions of the indicated histone mutant strains were spotted onto SC plates with 0 or 150 mM HU. (B) FLO8-HIS3 assay (see Materials and Methods) to measure cryptic transcription activation in the indicated histone mutants. Growth on SC-His media indicates the histone mutant strain activates a cryptic promoter within the FLO8 gene, leading to HIS3 expression. (C) The H2A E57A H2B M62E double mutant is lethal in yeast. WT or mutant histone genes are present on a LEU2 plasmid, and WT histone genes are on a URA3 plasmid that is shuffled out by 5-FOA selection. (D) Model of FACT binding to the H2A-H2B dimer. Our data suggest that FACT may target both a small hydrophobic patch near the DNA-binding interface that is comprised of H2B M62 and Y45 residues (highlighted blue) and residues H2A E57 and H2B L109 in the acidic patch (highlighted red). H2A-H2B structure was visualized from pdbID#1ID3 using PyMOL.
Our co-IP data indicate that H2A E57 and H2B M62 are important for FACT to bind free H2A-H2B dimers in vivo, as FACT binding to histone H2A and H2B was greatly diminished in these mutants relative to WT histones. However, unlike Spt16 or Pob3, these histone residues are not essential for yeast viability, likely because there is still considerable histone binding to FACT when the H2A E57A or H2B M62E mutants are expressed as the sole histone copy in yeast (i.e., in the absence of untagged WT histone; Figure S6, C and D, in File S1 and data not shown). We hypothesized that combining mutants in H2A E57 and H2B M62 might cause lethality in yeast by eliminating this residual FACT binding. Analysis of an H2A E57A H2B M62E double mutant indicated that it was lethal in yeast (Figure 5C), consistent with the hypothesis that these residues have redundant functions in FACT binding (Figure 5D).
Discussion
The FACT histone chaperone is essential for nucleosome assembly and disassembly in eukaryotic cells, in part through its interactions with histones H2A and H2B. However, the molecular mechanism by which FACT binds H2A and H2B and the histone residues important for FACT binding are not fully understood. While the nucleosome acidic patch is a common binding target of many chromatin-associated proteins, it has not been previously shown to be important for FACT binding to histone H2A and H2B. In this study, we show that specific residues in the nucleosome acidic patch (i.e., H2B L109 and H2A E57) are important for FACT binding to free H2A and H2B in yeast. We further show that acidic patch residues are important for histone occupancy in yeast, which in the case of H2B L109 could be due to its importance in FACT–histone interactions during nucleosome assembly.
The initial goal of this study was to characterize the functions of the four essential residues in yeast H2A and H2B (H2A Y58, E62, D91, and H2B L109), which are all located in the nucleosome acidic patch. Our data indicated that essential acidic patch residues are required for normal histone occupancy at a variety of genomic loci in yeast, even though these residues are located on the solvent-exposed nucleosome surface, and thus are unlikely to affect nucleosome stability. Our in-vitro studies indicated that the H2B L109 mutant does not significantly affect intrinsic nucleosome stability; similarly, a previous study has shown that histones lacking charged acidic patch residues (H2A E61A, E64A, D90A, and E92A in Xenopus) can form stable nucleosomes in vitro (Kim et al. 2015). Instead, our data indicate that H2B L109 is important for binding of the H2A-H2B dimer to FACT in vivo. While a defect in FACT binding could explain the decrease in histone occupancy observed for the H2B L109A mutant, it cannot explain the histone occupancy defects observed for the H2A D91A mutant, as this acidic patch residue does not affect histone binding to FACT or Nap1. We hypothesize that H2A D91 and potentially other essential acidic patch residues may be important for binding other chromatin assembly factor(s).
We show that a second acidic patch residue, H2A E57, is also important for FACT binding. Previously, it was reported that H2A E57 is important for histone occupancy and FACT recruitment at a number of loci in yeast (Cucinotta et al. 2015), although the mechanism underlying these effects was unclear. Our data indicate that H2A E57 (and H2B L109) are important for FACT to bind free H2A-H2B dimers in vivo. These data are supported by genetic evidence in yeast indicating that mutants in H2A E57 share a number of phenotypes with Spt16 and Pob3 mutants, including HU sensitivity and FLO8 cryptic transcription. Notably, we found that the effects of H2A E57A on FACT binding were similar in magnitude as a mutant in H2B M62. In-vitro studies have shown that H2B M62 is directly involved in binding the Spt16 C-terminal tail and abolishes FACT binding to H2A and H2B when mutated to glutamic acid (Kemble et al. 2015). However, our co-IP data indicate there is still considerable FACT binding to the H2B M62E mutant dimers in yeast in the absence of competing WT histones (Figure S6 in File S1). This observation can explain why the H2B M62E mutant is viable and grows normally, unlike spt16- or pob3-deletion mutants in yeast.
While our data indicate that some acidic patch residues were important for binding to FACT, none of the acidic patch residues tested was important for Nap1 binding to H2A and H2B. This is consistent with a recent crystal structure of Nap1 bound to H2A and H2B, as the H2A-H2B acidic patch is located distally from the Nap1–histone binding interface (Aguilar-Gurrieri et al. 2016). In contrast, H2A R77 in Xenopus (corresponding to H2A R78 in yeast) is located at the Nap1–histone binding interface, which can explain our data showing that the H2A R78A mutant causes a significant decrease in histone binding to Nap1 in vivo. These and other data indicate that Nap1 and FACT bind the H2A-H2B dimer through distinct binding interfaces.
In summary, our data indicate that residues in the histone acidic patch are important for normal histone occupancy in yeast, which may in part explain why they are essential for cell viability. This result is unexpected, as acidic patch residues have not previously linked to nucleosome assembly and do not appear to affect intrinsic nucleosome stability. Instead, we show that two acidic patch residues, H2B L109 and H2A E57, are important for binding of H2A-H2B dimers to the FACT histone chaperone in yeast. These findings suggest that FACT not only binds the small hydrophobic pocket comprised of H2B M62 and Y45, as has been previously reported (Kemble et al. 2015), but may also bind to residues in the histone acidic patch (Figure 5D). In support of this model, we show that an H2B M62E and H2A E57A double mutant, which eliminates both potential FACT binding sites, has a lethal phenotype in yeast, similar to spt16- or pob3-deletion mutants. While our co-IP and genetic data are consistent with this model, future studies will be required to test whether FACT directly binds to residues in the histone acidic patch or if acidic patch mutants affect FACT binding indirectly (e.g., by altering histone post-translational modifications or other chromatin-binding factors).
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.201939/-/DC1.
Acknowledgments
We are grateful to Peng Mao for helpful comments and suggestions. We thank Marian Laughery, Travis Ostbye, Marcos Perez, Abbey Porter, and Jacob Woodbury for expert technical assistance and helping construct plasmids and yeast strains. This work was supported by a grant from the National Institutes of Health (ES002614) from the National Institute of Environmental Health Sciences.
Footnotes
Communicating editor: M. Hampsey
Literature Cited
- Aguilar-Gurrieri C., Larabi A., Vinayachandran V., Patel N. A., Yen K., et al. , 2016. Structural evidence for Nap1-dependent H2A–H2B deposition and nucleosome assembly. EMBO J. 35: 1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain F. H., Yen Y. M., Masse J. E., Schultze P., Dieckmann T., et al. , 1999. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 18: 2563–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews A. J., Chen X., Zevin A., Stargell L. A., Luger K., 2010. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell 37: 834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache K. J., Garlick J. D., Canzio D., Narlikar G. J., Kingston R. E., 2011. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science 334: 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov N., Nourani A., Cote J., 2011. Histone chaperones: modulators of chromatin marks. Mol. Cell 41: 502–514. [DOI] [PubMed] [Google Scholar]
- Basnet H., Su X. B., Tan Y., Meisenhelder J., Merkurjev D., et al. , 2014. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature 516: 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R. J., Zhang Z., 2013. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 20: 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V., Chua G., Batada N. N., Landry C. R., Michnick S. W., et al. , 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6: e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta C. E., Young A. N., Klucevsek K. M., Arndt K. M., 2015. The nucleosome acidic patch regulates the H2B K123 monoubiquitylation cascade and transcription elongation in Saccharomyces cerevisiae. PLoS Genet. 11: e1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C., Tyler J. K., Churchill M. E., 2010. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem. Sci. 35: 476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltman M., Evrin C., De Piccoli G., Jones R. C., Edmondson R. D., et al. , 2013. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep. 3: 892–904. [DOI] [PubMed] [Google Scholar]
- Formosa T., 2012. The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 1819: 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., et al. , 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20: 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girish T. S., McGinty R. K., Tan S., 2016. Multivalent interactions by the Set8 histone methyltransferase with its nucleosome substrate. J. Mol. Biol. 428: 1531–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurard-Levin Z. A., Quivy J. P., Almouzni G., 2014. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 83: 487–517. [DOI] [PubMed] [Google Scholar]
- Hoch D. A., Stratton J. J., Gloss L. M., 2007. Protein-protein Forster resonance energy transfer analysis of nucleosome core particles containing H2A and H2A. Z. J. Mol. Biol. 371: 971–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A. J., Gallegos I. J., Laughery M. F., Meas R., Tran L., et al. , 2015. Histone sprocket arginine residues are important for gene expression, DNA repair, and cell viability in Saccharomyces cerevisiae. Genetics 200: 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondele M., Stuwe T., Hassler M., Halbach F., Bowman A., et al. , 2013. Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature 499: 111–114. [DOI] [PubMed] [Google Scholar]
- Jin Y., Rodriguez A. M., Stanton J. D., Kitazono A. A., Wyrick J. J., 2007. Simultaneous mutation of methylated lysine residues in histone H3 causes enhanced gene silencing, cell cycle defects, and cell lethality in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 6832–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalashnikova A. A., Porter-Goff M. E., Muthurajan U. M., Luger K., Hansen J. C., 2013. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface 10: 20121022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C. D., Laprade L., Winston F., 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099. [DOI] [PubMed] [Google Scholar]
- Kato H., van Ingen H., Zhou B. R., Feng H., Bustin M., et al. , 2011. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc. Natl. Acad. Sci. USA 108: 12283–12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble D. J., McCullough L. L., Whitby F. G., Formosa T., Hill C. P., 2015. FACT disrupts nucleosome structure by binding H2A–H2B with conserved peptide motifs. Mol. Cell 60: 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. A., Chatterjee N., Jennings M. J., Bartholomew B., Tan S., 2015. Extranucleosomal DNA enhances the activity of the LSD1/CoREST histone demethylase complex. Nucleic Acids Res. 43: 4868–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J., 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Makde R. D., England J. R., Yennawar H. P., Tan S., 2010. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467: 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P., Kyriss M. N., Hodges A. J., Duan M., Morris R. T., et al. , 2016. A basic domain in the histone H2B N-terminal tail is important for nucleosome assembly by FACT. Nucleic Acids Res. 44: 9142–9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse J. E., Wong B., Yen Y. M., Allain F. H., Johnson R. C., et al. , 2002. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 323: 263–284. [DOI] [PubMed] [Google Scholar]
- Matsubara K., Sano N., Umehara T., Horikoshi M., 2007. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells 12: 13–33. [DOI] [PubMed] [Google Scholar]
- McGinty R. K., Henrici R. C., Tan S., 2014. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514: 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. T., Haj-Yahya M., Ringel A. E., Bandi P., Brik A., et al. , 2016. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351: 725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S., Sanderson B. W., Delventhal K. M., Bradford W. D., Staehling-Hampton K., et al. , 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15: 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., Wu W. H., Lane W. S., Hampsey M., Reinberg D., 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400: 284–288. [DOI] [PubMed] [Google Scholar]
- Polo S. E., 2015. Reshaping chromatin after DNA damage: the choreography of histone proteins. J. Mol. Biol. 427: 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel L., Erard M., Cayrol C., Girard J. P., 2008. Molecular mimicry between IL-33 and KSHV for attachment to chromatin through the H2A–H2B acidic pocket. EMBO Rep. 9: 1006–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M., Noguchi S., Kawashima S., Okada Y., Enomoto T., et al. , 2009. Global analysis of mutual interaction surfaces of nucleosomes with comprehensive point mutants. Genes Cells 14: 1271–1330. [DOI] [PubMed] [Google Scholar]
- Wilkins B. J., Rall N. A., Ostwal Y., Kruitwagen T., Hiragami-Hamada K., et al. , 2014. A cascade of histone modifications induces chromatin condensation in mitosis. Science 343: 77–80. [DOI] [PubMed] [Google Scholar]
- Winkler D. D., Muthurajan U. M., Hieb A. R., Luger K., 2011. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 286: 41883–41892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick J. J., Kyriss M. N., Davis W. B., 2012. Ascending the nucleosome face: recognition and function of structured domains in the histone H2A–H2B dimer. Biochim. Biophys. Acta 1819: 892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Yeast strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.