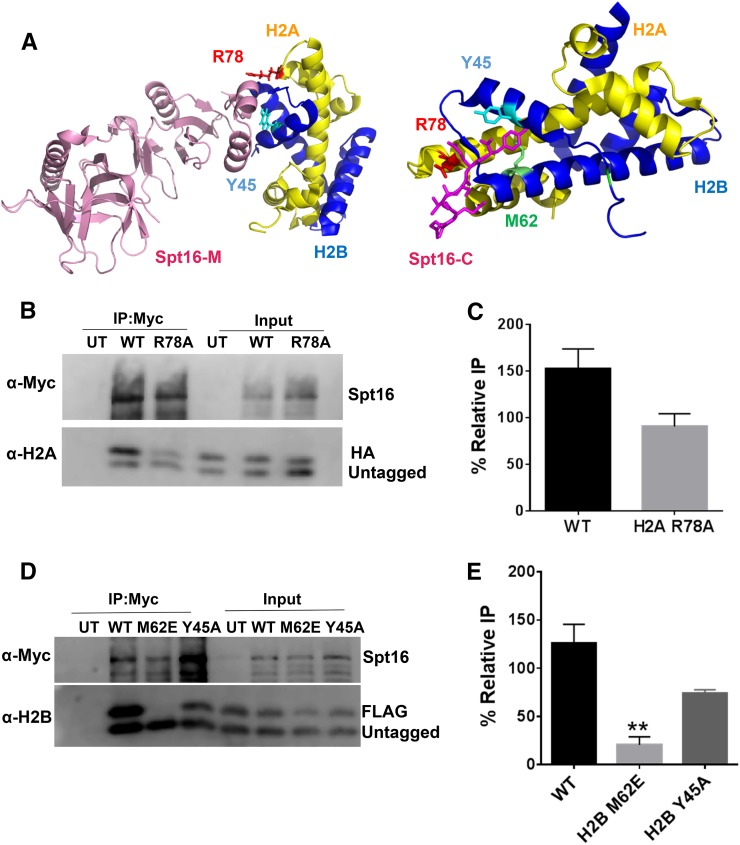
Figure 4.
Analysis of FACT binding to histone mutants identified from structural studies. (A) Published crystal structures of FACT in complex with histones pdbID#4KHA (left) and pdbID#4WNN (right). Residues identified as important for FACT binding (e.g., H2A R78, H2B Y45, and H2B M62) are indicated. Image was created using PyMOL. (B and C) H2A R78A does not inhibit binding to FACT. (B) Co-IP analysis using anti-myc beads of nuclease-treated Spt16-myc yeast whole-cell extracts containing HA-tagged WT or H2A R78A mutant histones. An untagged WT is coexpressed. (C) Quantification of the percent IP of the tagged band relative to the untagged band. Graphs represent the mean and SEM of at least three independent replicates. (D and E) H2B M62E significantly inhibits FACT binding. (D) Co-IP analysis using anti-myc beads of yeast whole-cell extracts containing myc-tagged Spt16 and FLAG-tagged WT H2B, H2B M62E, or H2B Y45A mutant histones coexpressed with untagged WT histones. (E) Quantification of the percent IP of the tagged band relative to the untagged band. Graphs represent the mean and SEM of at least three independent replicates. ** P < 0.01.