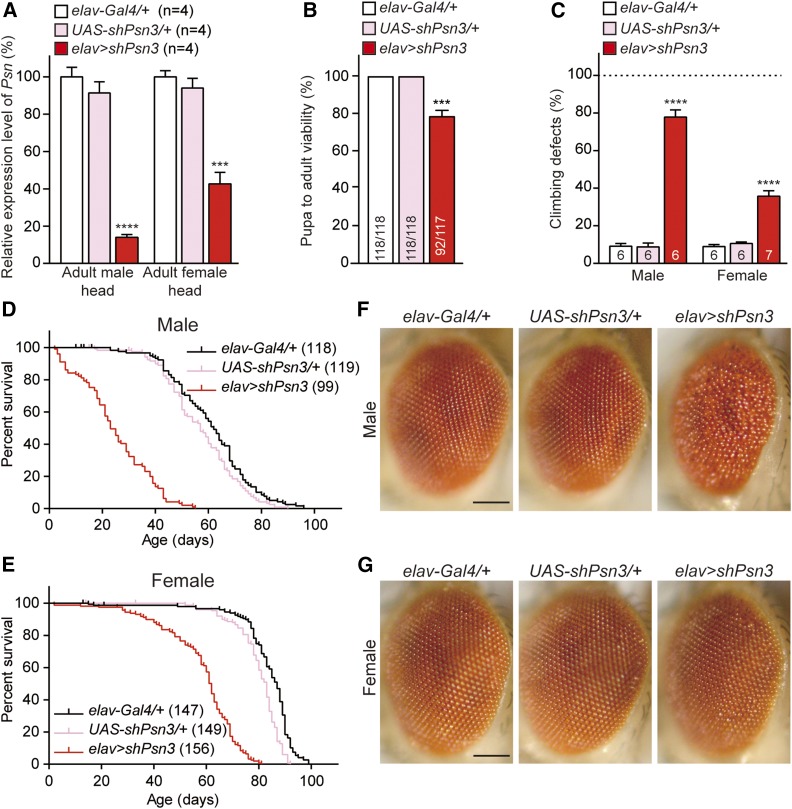
Figure 4.
Similar but less severe phenotypes associated with a different Psn shRNA line. (A) Neuron-specific Psn KD using a different shRNA line results in significant reduction of Psn mRNA levels in 3-day-old adult heads of elav > shPsn3 males (83 ± 3.2%) and females (54 ± 2.8%), compared to controls. qRT-PCR analysis of Psn mRNA level in adult fly heads expressing shPsn3. Psn mRNA levels are normalized to rp49 mRNA levels as internal control. Total RNA was extracted from 20 adult heads per genotype. n = 4 independent experiments. (B) Neuron-specific Psn KD in elav > shPsn3 reduces pupa-to-adult viability (92/117). Viability was calculated by dividing the total number of flies by the total number of pupae, and shown as the percentage of pupae surviving to adulthood. n = 6 independent experiments, ≥110 flies per genotype (∼20 flies per experiment). (C) elav > shPsn3 causes defects in climbing ability in both males and females. Bar indicates percentage of failed climbers. Age = 3 days, n = 6 independent experiments, ≥110 flies per genotype (∼20 flies per experiment). (D, E) elav > shPsn3 causes severe mortality in male (D) and female (E) flies. Survival of Gal4 controls (elav-Gal4/+, black), UAS controls (UAS-shPsn3/+, pink), and elav > shPsn3 flies (elav-Gal4/+;; UAS-shPsn3/+, red). Lifespans were plotted using the Kaplan-Meir method. (F) elav > shPsn3 male flies exhibit severe rough eye phenotypes. (G) In contrast to females with a stronger shPsn2 line (elav > shPsn2), females of elav > shPsn3 show normal compound eyes. Representative images showing adult eye of controls and elav > shPsn3 flies. Bar, 0.1 mm. All data are expressed as mean ± SEM. Statistical analysis was performed using one-way ANOVA with Dunnett’s post hoc comparisons. ***P < 0.001, ****P < 0.0001.