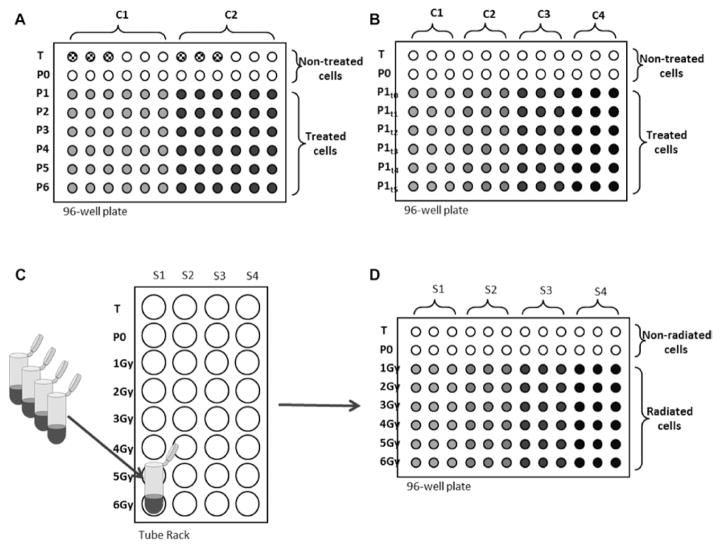
Figure 3.
Schematic representation of three different experimental layouts for analysis of DNA strand breaks using the automated FADU assay. (A) Cells attached to a 96-well plate are treated with two different compounds (C1 and C2) in six different concentrations (P1–P6). T and P0 are untreated controls; P0 represents the endogenous amount of DNA strand breaks and T the total DNA amount. In this example, three wells of T values are designated to assess the interference of compounds or solvents used with the fluorescence signal. For this purpose, cells are treated with the compound and/or solvents and compared to the untreated T values. This layout allows six replicates for conditions P0–P6 and three replicates for treated T and untreated T values, respectively. (B) Cells attached to a 96-well plate are treated with four different compounds (C1–C4) in one single concentration (P1). After damage occurs (P1t0), cells were incubated for different repair times (P1t1–Pt5). T and P0 are untreated controls; P0 represents the endogenous amount of DNA strand breaks and T the total DNA amount. This layout allows three replicates for each condition. (C) Lymphocytes from four different subjects (S1–S4) are irradiated in 2 ml tubes with doses ranging from 1 to 6 Gy. Cooled tube rack is placed in the liquid handling device (LHD) workspace and (D) samples are transferred to a pre-cooled 96-well plate. T and P0 represent non-radiated cells. This layout allows three replicates for each condition.