Abstract
Cellular plasma membranes are laterally heterogeneous, featuring a variety of distinct subcompartments that differ in their biophysical properties and composition. A large body of research has focused on understanding the basis for this heterogeneity and its physiological relevance. The membrane raft hypothesis formalized a physicochemical principle for a subtype of such lateral membrane heterogeneity, wherein the preferential associations of cholesterol and saturated lipids drives the formation of relatively packed (ordered) membrane domains that selectively recruit certain lipids and proteins. Recent years have yielded new insights into this concept and its in vivo relevance, primarily owing to the development of biochemical and biophysical technologies.
Introduction
Only one year after the seminal paper of Singer and Nicholson proposed the fluid-mosaic model for biomembrane organization1, the first observations that cell membranes can be separated into detergent-labile and detergent-resistant fractions2 sparked the idea that distinct membrane sub-populations are present in biological membranes (for a brief history of biomembrane models, please see ref3). This finding was followed by a number of observations suggesting cellular membranes are laterally heterogeneous at the sub-micron level4–9. The membrane raft (or lipid raft) hypothesis was a specific formulation of the broad concept of lateral membrane inhomogeneity, proposing that the interactions between certain lipids (e.g., cholesterol, relatively saturated lipids, glycosylated lipids) in the plane of the membrane drive the formation of functionally important, relatively ordered membrane regions that recruit other lipids and proteins10. This concept was supported by observations of biomimetic model membranes, where there is clear evidence that certain lipids interact preferentially with one another, engage in collective behaviour, and generate large scale lateral domains as a consequence of liquid–liquid phase separation11.
However, the in vivo presence and relevance of such ordered membrane domains was unclear, due in part to the lack of direct observations of these domains and uncertain definitions of the lipid raft concept. To address this uncertainty, a consensus operational definition of “lipid rafts” was formulated in 2006, with available evidence suggesting that rafts are heterogeneous, dynamic (in terms of both lateral mobility and association-dissociation), cholesterol and sphingolipid enriched membrane nano-domains (10–200 nm) that have the potential to form microscopic domains (>300 nm) upon clustering induced by protein–protein and protein– lipid interactions12 (Fig. 1). These domains are present both in the inner and outer leaflets of an asymmetric cell membrane, are presumably coupled across leaflets13,14, and form functional platforms for regulation of cellular functions15. Recently, a number of emerging biochemical and biophysical techniques have provided support for the presence of such domains in cells and suggested key roles for membrane heterogeneity in various cellular functions. The conservation of lipid rafts in the tree of life has also been demonstrated (Supplementary Box S1), providing further support for their biological significance. However, lipid rafts continue to escape direct microscopic detection, thus the presence and exact nature of rafts in live cells remains debated, particularly as different methodologies can often yield seemingly contradictory results16.
Figure 1. General overview of lateral heterogeneity in the plasma membrane.
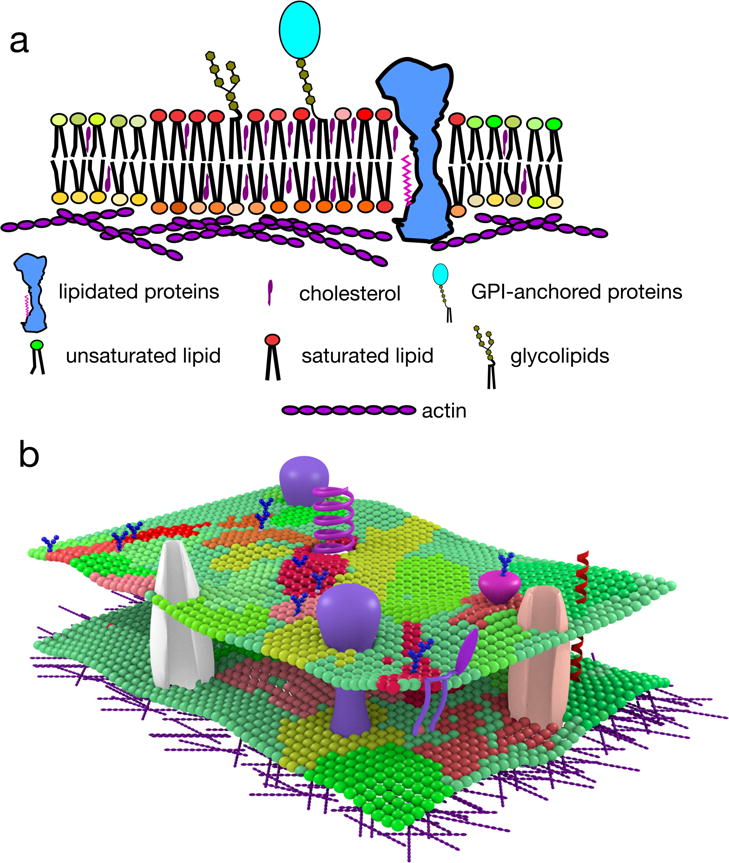
a | Lipid raft domains are small, highly dynamic and transient plasma membrane entities enriched in saturated phosho-, sphingo- and glycolipids, cholesterol, lipidated proteins and glycosylphosphatidylinositol (GPI)-anchored proteins. Enrichment in these hydrophobic components endows lipid rafts with distinct physical properties, including increased lipid packing and order, and decreased fluidity. In addition to membrane components, cortical actin plays an active role in domain maintenance and remodelling. Further, membrane lipids are asymmetrically distributed in the inner and outer leaflets, and this may further impact membrane organization. b | It is likely that membrane organization is not binary (i.e. highly specified raft and non-raft regions), but rather consists of various raft-like and non-raft domains with distinct compositions and properties.
Here, we define rafts as transient, relatively ordered membrane domains, whose formation is driven by lipid– lipid interactions, and discuss the technological advances that have reignited the excitement around this concept and its in vivo relevance. In particular, we focus on the current understanding of the mechanisms of raft formation and maintenance and conclude with a discussion of the challenges remaining in this dynamic field.
Studying lipid rafts
The definition of rafts has been, in large part, influenced by the development of methodologies available for their investigation. The term “lipid rafts” has been generically applied to many distinct, though potentially related, types of membrane assemblies (Fig. 2a). The techniques and tools to visualize and study membrane heterogeneity have evolved considerably since the introduction of the concept (Fig. 2b–d), with the recent advent of super-resolution optical microscopy (Supplementary Box S2) providing a key tool towards potentially resolving the continuing controversy.
Figure 2. Tools to study membrane domain organization, composition and function.
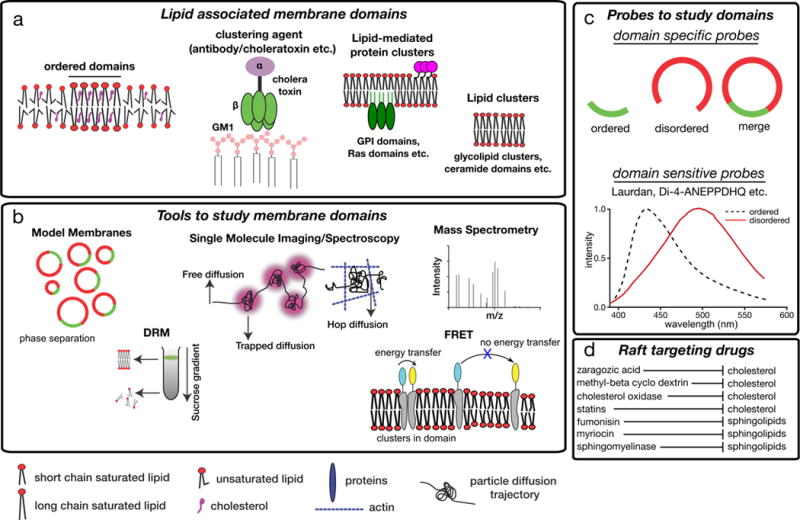
a | In principle, membrane domains can be pure lipid clusters, but in most physiologically-relevant cases, also involve proteins, including glycosylphosphatidylinositol (GPI)-anchored protein clusters or clusters of Ras proteins. These domains can be purely lipid-driven entities, such as domains established through liquid–liquid phase separation in model membranes. They can also be induced by clustering agents, such as cholera toxin which binds to monosialotetrahexosylganglioside (GM1) or by antibodies recognizing surface receptors b | Tools that are commonly used to investigate membrane domains. These include various model membranes (such as synthetic giant unilamellar vesicles (GUVs) and cell-derived giant plasma membrane vesicles (GPMVs)); detergent resistance assays, wherein raft-like membrane regions persist as detergent-resistant membranes (DRMs), whereas non-raft components are fully solubilized; single molecule imaging to evaluate the diffusion of membrane molecules; fluorescence spectroscopy (such as Förster resonance energy transfer (FRET)), and mass spectrometry. c | Also various probes can be used to study raft domains. Domain selective probes partition selectively to one of the domains while domain sensitive probes partition to both domains and change their photophysical behaviour (for instance absorbance and emission spectra) depending on the nature of the surrounding lipid environment. d | Treatments that interfere with cholesterol or sphingolipid levels have been used to disturb rafts in cells and can shed light on the cellular functions of these domains.
Biochemical tools
The first evidence for a laterally heterogeneous cell membrane was the observation of differential solubilisation of membrane lipids and proteins by detergents in the 1970s2. The basis of the assay is that non-ionic detergents (under certain conditions, most notably cold temperatures) fractionate cellular membranes into detergent-soluble membrane fractions (DSMs) and detergent-resistant membrane fractions (DRMs) (Fig. 2b). These fractions have clearly distinct compositions, with DRMs enriched in cholesterol, sphingolipids17,18, and glycophosphatidylinositol (GPI)-anchored proteins5. Although extraction of DRMs became the method of choice for probing membrane raft compositions, it quickly became clear that they cannot reflect the native composition and organization of lipid rafts in living cells. For example, the protein composition of DRMs varies widely depending on the choice of detergent used for isolation19. Similarly, subtle variations in temperature or detergent concentration yield different results, and considerably modify the organization of membrane proteins20, which has led to contradicting reports of protein composition of rafts. Thus, while DRMs may inform on the propensity of some molecules to associate with specialized membrane regions21,22, they do not faithfully reflect the native molecular or biophysical context of rafts23 and therefore require confirmation by more robust and consistent methods, such as those discussed in the remainder of this section (for an excellent recent example, see ref 22).
Biophysical tools
In parallel with studies of DRMs isolated from cells, artificial membrane models have been developed and used to study the liquid–liquid phase separation believed to underlie the physical principle behind lipid raft formation24 (Fig. 2b). Across various experimental set-ups, membranes consisting of relatively saturated lipids with a high melting temperature, unsaturated phospholipid species with a low melting temperature and cholesterol can separate into two distinct liquid phases: a relatively packed and ordered phase enriched in saturated lipid species and cholesterol25 (termed the liquid-ordered, or Lo), and a more fluid disordered phase comprising mainly the unsaturated lipids26,27 (termed the liquid-disordered, or Ld). Owing to its high molecular packing and enrichment of sterol and saturated lipids, the Lo phase is considered as the model for lipid rafts. Biomimetic monolayers28, supported lipid bilayers29, nanoscopic bilayers vesicles30, and giant unilamellar vesicles (GUVs)26 have all been used to elucidate the molecular details behind this phase separation31,32; however, despite their important role in revealing the physical principles of liquid-ordered domain formation, a number of caveats and limitations prevent direct translation of findings from model membranes to biological ones. First, most such experiments are performed in lipid-only systems, and although there are methods for incorporating integral membrane proteins into artificial systems33,34, they are complex and inefficient, and very rarely result in high protein-to-lipid ratios. This is in contrast to biological membranes, in which proteins are estimated to constitute up to 25% of the membrane’s cross-sectional area35. Perhaps because of the dearth of proteins, some features of domains established in synthetic membranes may not be representative of biology. For example, ordered domains in synthetic membranes have extremely high molecular order and tight packing, whereas the other extreme is observed in the disordered domains27,36. These caveats can be avoided by studying more natural systems such as giant plasma membrane vesicles (GPMVs)37,38. GPMVs are cell-derived, intact plasma membrane vesicles that maintain the lipid39 and protein40 diversity of the cellular membrane, with the notable exceptions of an assembled cortical actin cytoskeleton, phosphorylated lipids41 and strict lipid asymmetry between separate leaflets of the membrane bilayer (we refer readers to ref42 for a detailed discussion of the advantages, caveats, and applications of GPMVs). In these biological membranes, the contrast in molecular order between coexisting ordered and disordered domains is much smaller than, for example, in synthetic GUVs. These differences in molecular order between the two phases may account for the fact that GUV ordered phases exclude nearly all transmembrane proteins and most fluorescent lipid probes (see below), whereas the same molecules are sometimes enriched in the ordered phase in GPMVs (as would be expected for lipid rafts in vivo)36,43 (see also BOX 1).
Box 1. Model membranes to study formation and organization of lateral domains.
Combining a relatively saturated lipid, an unsaturated lipid and cholesterol in a model membrane, often results in liquid–liquid phase separation and the establishment of two distinct phases (still liquid in nature)11. One of these phases (liquid-ordered (Lo)) is more viscous that the other (liquid disordered (Ld)) owing to the tighter packing and higher molecular order of its constituent lipids91. This Lo phase is believed to be analogous to lipid rafts in cellular membranes.
Supported lipid bilayers (SLBs, see Figure panel a) are planar bilayers formed on glass or mica surfaces29. As these membranes are planar, they are highly amenable to microscopic imaging, either by light or Atomic Force Microscopy, allowing observations of the topology of nanodomains that are not resolvable with diffraction-limited optical microscopy (Supplementary Box S2). The artefacts caused by the solid support in SLBs are avoided by the use of free-standing membranes like giant unilamellar vesicles (GUVs) (see Figure panel b), which have been frequently used to investigate domain dynamics and morphologies187. The limitation of synthetic model systems is their simple composition, which does not fully recapitulate that of the cell membrane.
Giant plasma membrane vesicles (GPMVs) are obtained from cell membranes37. Like GUVs, these form micron-scale lateral liquid domains (confirming the capacity for liquid–liquid phase separation in biological membranes), but do so while maintaining the broad compositional features of the native plasma membrane. The most notable differences between GUVs and GPMVs are lipid complexity and the presence of abundant transmembrane proteins37,38, incorporation of which into SLBs and GUVs remains technically challenging. The biophysical properties of GPMVs are somewhat distinct compared to artificial membranes37,92,188. For example, the difference in packing between Lo and Ld domains in GPMVs is much smaller than in GUVs (Figure Box panel c; generalized polarization is a relative index of lipid packing, with +1 representing maximally ordered and -1 representing maximally disordered membranes), which may explain why transmembrane proteins can associate with Lo phase in GPMVs43 but not in GUVs189. Despite these differences, most of the core features of coexisting liquid-ordered and -disordered domains in these model systems are fundamentally similar24.
Analytical tools
In cells, rafts are believed to be nanoscopic (<200 nm)7,8, and they are therefore unresolvable using conventional optical microscopy with its ≈250 nm resolution limit set by diffraction (see Supplementary Box S2). Although confocal microscopy studies have reported co-localization of certain molecules with putative lipid raft markers (such as cholera toxin) as evidence of their raft association44, in general the resolution of such systems is insufficient to directly assay raft domain structure and composition. To overcome this limitation, several optical tools have recently been developed45,46 and applied to investigate nano-scale structures and dynamics in cells. For instance, super-resolution optical microscopy approaches such as photoactivated localization microscopy (PALM), stimulated emission depletion (STED) microscopy (Supplementary Box S2), and near-field scanning optical microscopy (NSOM) have been used to visualize lipid-mediated protein clustering47–50.
For more dynamic measurements, single-molecule based techniques such as single particle tracking (SPT) have been used to evaluate the diffusion of membrane molecules and relate them to concepts of heterogeneous organization of the membrane51. Such studies can reveal oligomerization52, transient arrest, domain incorporation, and/or confined and hop (compartmentalised) diffusion53 of tracked molecules (Fig. 2b). Recently interferometric scattering (iSCAT) microscopy has further increased the sensitivity of SPT54, and shown great potential for assessing membrane heterogeneity. For example, iSCAT was used to show that lipids can transiently stall and incorporate into sub-20nm domains within model membranes55,56. Complementary to SPT, fluorescence correlation spectroscopy (FCS) has been applied in combination with spot variation (svFCS57) or a STED microscope (STED-FCS58) to probe lateral diffusion of membrane components over various length scales. Particularly in STED-FCS the size of the observation spot can be reduced tô20–40nm, revealing underlying nanoscopic features of the plasma membrane57–59. Finally, Förster resonance energy transfer (FRET, Fig. 2b) is a key tool for investigating membrane raft structure and composition60,61. The spatial regime probed by this technique makes it ideal for studying nanoscopic domains, and it has been applied to both model membranes62 and live cells63 not only to probe the existence of domains, but also to define their size62,64 by using fluorescent probes with different FRET efficiencies. For a detailed review on these techniques and their caveats, see ref45.
Most of the above-mentioned methodologies rely on fluorescent labels. This is a particular issue in investigation of membranes because the behaviour of lipids is inherently dependent on their amphiphilic properties and molecular packing, both of which are potentially affected by tags such as fluorophores, which are often nearly the size of the lipid molecules. Thus, the native behaviour of lipids is often greatly altered by the reporter36. To address this concern, a number of label-free techniques have been developed. Mass spectroscopy (Fig. 2b), for instance, is one the most accurate tools to address the lipid and protein composition of membranes without the necessity of external labelling65, and has been used for label-free determination of membrane domain composition in model and cell-derived membranes66–71. iSCAT microscopy has also facilitated label-free observation of the dynamics of ordered domains in model membranes72. Raman spectroscopy is another label-free technique that has been successfully applied to monitor membrane domain composition73. Likewise, small-angle neutron scattering has also been used to detect raft-like domains74 and their size75. Finally, electron microscopy has the necessary resolution for obtaining a snapshot of molecular arrangements at the cell surface, and a number of studies of outer and inner leaflet lipid tethered proteins, including GPI-anchored proteins, glycolipids and Ras proteins, have revealed the nanoscopic organization of proteins in rafts76. One potential caveat of these methods is that they usually require cell fixation and staining, which are notoriously problematic in visualizing fluid lipid molecules. Therefore, fluorescence microscopy remains a preferred technique for direct live imaging of the putative lipid raft components, and this necessitates continued optimization of fluorescent labels for membrane components.
Probes selective to membrane domains
Non-perturbing, specific labelling of raft or non-raft domains in cells has been, and remains, one of the foremost challenges of the field. Several fluorescent markers have been used to distinguish between different membrane compartments, including cyanine dyes (e.g., DiO, DiI, DiD)77, polycyclic aromatic hydrocarbons (e.g. naphtopyrene)78, and fluorescently-labelled lipids36,79 (Fig. 2c). As mentioned above, the reliability of these fluorescent lipid analogues depends strongly on the choice of both the native lipid and the fluorescent moiety36. The least perturbing fluorescent lipids are the intrinsically fluorescent cholesterol analogues such as dehydroergosterol80 and cholestatrienol81; however, their poor photophysical characteristics in comparison with artificially tagged lipids have prevented their wide application. In the case of phospholipids, it is often challenging to preserve the natural physicochemical behaviour of the lipid after attaching a fluorophore82,83. In general, the least perturbing strategy is labelling the headgroup rather than the acyl chain, and adding a hydrophilic linker to ensure that the fluorophores (which are often membrane active) do not perturb the headgroups of the surrounding lipids84.
In addition to lipid analogues that can reveal general organization of the membrane into subdomains of variable composition, reporters that selectively bind core raft components can potentially be employed to visualize domains. These include cholesterol binding agents such as filippin85 and Perfringolysin O86, sphingolipid reporters such as ostreolysin A87, lysenin88 and pleurotolysin89 as well as ganglioside lipid ligands such as cholera toxin90. The major caveats for these probes are (a) their potential perturbation of native membrane organization, for example by inducing clustering of their binding partners, as is the case for cholera toxin; and (b) their reduced specificity in the cellular context where they can potentially bind off-target species, thereby lowering their specificity for raft domains.
Probes sensitive to membrane environments
Coexisting lipid domains inherently have different physico-chemical properties. A particular property that defines lipid rafts is their tight lipid packing owing to the condensing interactions between relatively saturated lipids and cholesterol91. Notably, there is not a specific, unique molecular packing that would be common to the plasma membranes and their domains in different cells and contexts92. The diversity in membrane compositions and physical properties across cell types, and within cell types during physiological events like secretory granule secretion92 or cell cycle progression93, implies that a wide spectrum of different packing states exists in living cells. This lipid packing can be quantified using probes such as Laurdan that sense the level of hydration in the bilayer94 in combination with two-photon32 or conventional confocal microscopy95. These probes shift their emission spectra depending on the polarity (i.e. aqueous content or hydration) of the environment96 (Fig. 2c), providing a ratiometric, concentration-independent quantification of the local environment, which for membranes is determined largely by lipid packing97 (i.e. more tightly packed membrane exclude water more efficiently). Imaging of membrane packing using these probes has been applied to investigate membrane heterogeneity in live cells47,98. More recently, in addition to spectral shift, the lifetime99 and energy-transfer100 properties of these probes have been used to further investigate lipid packing in living membranes, expanding the toolbox and sensitivity of their potential applications. An important future development will be to enable efficient use of these probes in super-resolution microscopy.
Raft targeting drugs
A common paradigm to study the physiological roles of lipid rafts has been to use drugs or enzymes that impair the structure and function of these domains (Fig. 2d). Since cholesterol is usually thought to be enriched in rafts, the most common raft disrupting agent is methyl-beta cyclodextrin (MBCD), which selectively and efficiently removes cholesterol from membranes101. However, it needs to be considered that MBCD-mediated cholesterol removal has a broad pleitropic effects beyond raft disruption. For example, it increases membrane permeability to ions and thereby disrupts membrane potential102, and potentially is cytotoxic103. Moreover, this reagent appears to preferentially remove cholesterol from liquid-disordered (non-raft) domains in model membranes104, which can induce unexpected and inconsistent21,67 effects on lipid packing in more complex membranes. Drugs targeting cholesterol synthesis (statins105 and zaragozic acid106) or cholesterol-modifying enzymes (e.g. cholesterol oxidase107) have the potential to replace MBCD for raft disruption, but their specificity and effectiveness remain to be conclusively demonstrated. Other core components of rafts in cells are sphingolipids, and a number of reagents can interfere with their synthesis (e.g. fumonisin108 or myriocin109) or stability (e.g. sphingomyelinase110), though these suffer from potential off-target effects, such as the impact on general sphingolipid metabolism and the generation of ceramides, which then alter membrane properties in a different way.
Molecular dynamics simulations
One of the biggest remaining challenges in understanding biomembranes is how the myriad of interactions between membrane molecules determines membrane organization. Overcoming this challenge requires a combination of complementary experimental approaches, as well as in silico techniques that integrate results from experiments (e.g. data about the structure and energetics of the system) into a simulation framework that ideally reconstitutes the natural behaviour in silico based solely on physical interactions111. An inherent advantage of such in silico approaches is that they model a multitude of molecules simultaneously with high spatial (atomistic level) and temporal (ns-μs) resolution, without relying on exogenous probes or labels. Thus, in silico molecular dynamics simulations can be regarded as a computational microscope112, capable of visualizing molecular behaviour with unprecedented precision. Currently, such computational microscopes have the inverse limitations of optical microscopes, in that they reveal only fast (microseconds) and nanoscopic (thousands of molecules) processes, as opposed to the slow and large scales accessible by optical microscopy112,113. To close the gap between computational and experimental approaches, methods such as coarse graining have been developed to extend the spatiotemporal scale of molecular dynamics simulations without sacrificing the molecular details114. Such simulations have been successfully employed to study lipid–lipid and lipid–protein interactions115,116 and lipid domains in complex membranes14,117,118. It is important to note that such in silico observations are inherently model-driven and must be ultimately verified by experiments. Unfortunately, in the case of membrane domains, the spatiotemporal gap between direct observables of simulations and experiments is still too large to allow direct comparisons111. However, efforts from both directions are aiming to bridge this divide towards a molecular understanding of how complex membrane components self-organize into functional substructures.
Nature and composition of lipid rafts
Dissecting the physical properties – lifetime, size, and area coverage – of lipid rafts in the cellular environment remains one of most vexing issues in the field. Computational models confirm the intuitive assumption that both temporal and spatial compartmentalization of membranes into domains is crucial for membrane function119. Unfortunately, both the small size and short lifetime of putative raft domains in vivo complicate direct measurement of their properties in living cells. Furthermore, the complexity of plasma membranes suggests that a spectrum of raft-like domains, with varying sizes and lifetimes can be established in vivo92,98,120, further complicating interpretations of experimental measurements. The original model of lipid rafts suggested the existence of a liquid disordered (non-raft) membrane punctuated by more ordered (raft) domains of minimal coverage121. However recent data indicate a much greater contribution of ordered raft-like regions, suggesting that ordered membrane domains might be in fact dominating, possibly covering the majority of the plasma membrane, with interspersed less-ordered (non-raft) domains47,59 (Fig. 3a). Both the area fraction as well as the size and lifetime of membrane domains may be further tuned by cellular processes such as signalling and membrane trafficking (Fig. 3b) which makes it even more challenging to draw conclusions regarding these membrane domains.
Figure 3. Area coverage of membrane domain and domain size.
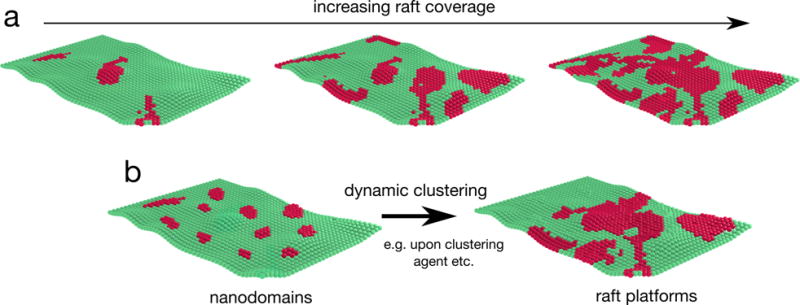
a | Models of membranes with varying raft coverage. Total raft coverage in a given membrane may vary broadly, ranging from small isolated domains to percolating raft phases of increasing size. The specific organizational state depends on a variety of factors, including cell type, specific cellular conditions (e.g. cell cycle), and/or the identity of the membrane (e.g. plasma membrane versus intracellular membranes). b | Another mode of modulation of membrane organization can occur without changing overall raft abundance. For example, the size and/or lifetime of individual domains may be influenced by cellular processes such as endo- and exo-cytosis, lipid metabolism, etc. In addition, binding of clustering agents (antibodies and toxins) to their receptors can promote the formation of large membrane domains.
In the original formulation, lipid raft formation was based on preferential interactions between sphingolipids and cholesterol10. Consistent with this notion, sphingomyelin has been identified as a core component of detergent resistant membranes2 and ordered lipid phases122, in part due to strong hydrogen-bonding interactions with cholesterol123,124 (Fig. 4a). However, the relative partitioning of cholesterol between more and less ordered domains is less clear, with experimental30 and computational125 studies suggesting that it is present in high abundance in both ordered (raft-like) and disordered (non-raft) phases, though with an enrichment in more ordered domains. Ganglioside lipids were also found to interact with cholesterol and form cholesterol rich domains in model membranes70, and these lipids have been consistently detected in the ordered domains of model membranes90. In addition, other lipids such as relatively saturated phospholipids have often been associated with raft-like environments, especially in model membranes.
Figure 4. Regulation of membrane domains.
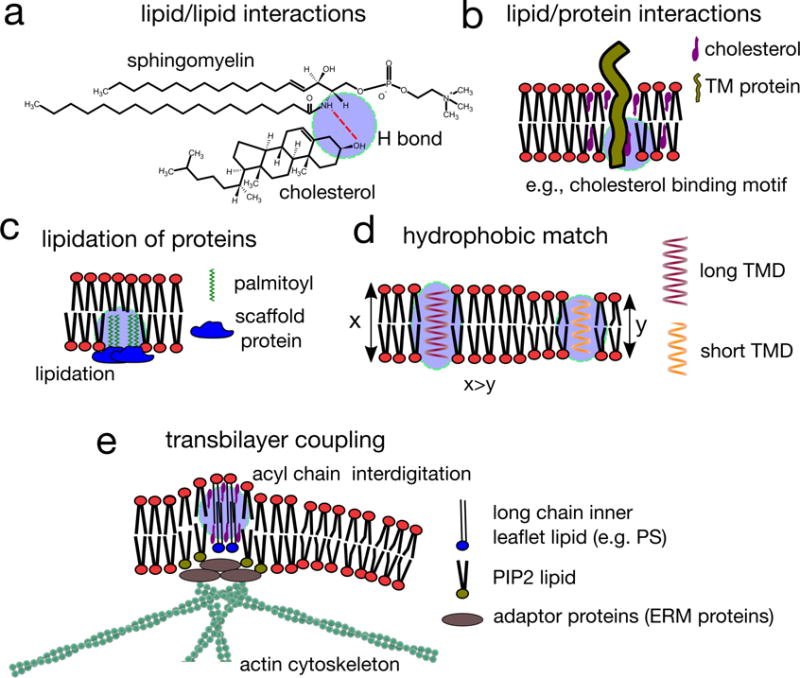
a | Lipid–lipid interactions, in particular interactions between cholesterol and sphingolipids (but also between other relatively saturated lipids) are the defining feature of lipid-driven ordered domain formation. The preferential interaction between sphingolipids and sterols is due to the saturation of sphingolipid hydrophobic tails, but also hydrogen bonding between these lipid species. The amide of the sphingolipid backbone can both donate and accept a hydrogen bond, and these hydrogen bonds are within the interfacial region of the membrane, where the relative paucity of water increases the relative stability of these bonds. b | Some proteins harbour lipid binding domains, interacting with cholesterol or sphingolipids, and these lipid–protein interactions may determine the affinity of proteins for ordered lipid domains. c | Lipidated proteins, modified by the attachment of a saturated acyl chain (such as palmitoyl moieties), are recruited to raft domains, but may also nucleate and recruit membrane domains if they are intergrated into a relatively static protein scaffold. d | Hydrophobic interactions can contribute to membrane domain organization and composition. In particular, proteins possessing transmembrane domains (TMD) of different lengths prefer different lipid environments to protect their hydrophobic TMDs from exposure to the aqueous surrounding. For example, proteins with long TMDs were found to associate with domains harbouring long chain saturated lipids (top). When there is a mismatch between the length of the TMD and the local lipid environment in which the protein resides, protein–protein interaction might be favoured instead, leading to local protein concentration (bottom). e | The immobilization of inner leaflet lipids containing long saturated acyl chains by actin clusters results in the engagement of long acyl chain containing lipid-anchored proteins (such as glycosylphosphatidylinositol (GPI)-anchored proteins) located in the outer leaflet, which, in the presence of cholesterol, induces their active clustering. This results in locally ordered, transbilayer nanodomains, which are dynamic owing to the dynamics of actin clusters, and may form even in conditions that do not favour liquid–liquid phase separation of lipids or other supporting interactions.
Whereas the biophysical basis for the lipid composition of rafts can be explained by these simple principles, the fundamental bases of selective incorporation of proteins into raft domains remains largely mysterious. In general, proteins interacting with the membrane via lipid anchors follow the rules set by the lipids: saturated lipid anchors such as GPI or palmitoyl moieties generally favour ordered membrane environments, while branched or unsaturated anchors like prenyl groups are non-raft preferring126. In fact, GPI-anchored proteins were some of the first proteins identified in DRMs5 and later in the ordered domains of model membranes33,127. Lateral GPI-anchored proteins domains have been extensively characterized by single molecule approaches128,129. Although their relationship to membrane rafts remains unresolved130, the membrane-mediated interactions between these lipid anchored proteins and lipids almost certainly regulate membrane structure and function22,69.
However, lipidated proteins are certainly not the sole protein species associating with rafts. In fact, in a recent experiment, 35% of all plasma membrane proteins were found in ordered domains of GPMVs43. These ‘raftophilic’ proteins included GPI-anchored proteins and palmitoylated proteins as expected (both contributing to one-third of the identified proteins)43. However, the remaining one-third of raft-associated proteins contained neither GPI nor palmitoyl anchors and the mechanisms of raft association of many of these proteins are currently unclear. Some proteins are known to become more raftophilic upon oligomerization, which may modulate their activity131. Recently, a database of putatively raft-associated proteins has been established based on mass spectrometry studies of isolated DRMs (RaftProt)132, though it is important to emphasize that these may be subject to the DRM-associated artefacts described above. As the actual protein content of membrane domains is uncertain, few generalizable insights into the structural determinants of raft partitioning for transmembrane proteins are available133. Interestingly, a recent study demonstrated that the length of the transmembrane domain (TMD) appears to be a key feature determining the raftophilic properties of a protein, with longer TMDs preferentially targeting the protein to the thicker ordered domains134.
Mechanisms of domain regulation
Although the raft concept and its in vivo relevance have been controversial, the principle of lateral membrane compartmentalization by lipids is intuitive: there are clear differences in the interaction affinities between various lipids, and these differences may be sufficient to produce a heterogeneous lipid distribution. For systems in thermodynamic equilibrium (including synthetic and biological model membranes42), the manifestation of these phenomena is macroscopic phase separation, which can be regulated by temperature135, composition21,26,67, or by specific interactions that enhance the inherent connectivity of separating components resulting in enhanced clustering136. However, cell membranes in vivo are not closed systems in chemical and thermodynamic equilibrium, and many potential regulatory modes contribute to the ultimate output of the inherent self-organizing capacity of biological lipids and their separation into distinct domains (Fig. 4).
Lipid-lipid and lipid-protein interactions
In the traditional raft model, the formation of raft domains is driven mainly by the preferential binding of cholesterol to sphingolipids124 and possibly other lipids such as gangliosides69 (Fig. 4a). However, an inherent limitation of studying the factors that regulate raft domain properties is the difficulty of measuring those properties in situ. To address this limitation, several recent reports93,137 have focused on factors that regulate the temperature at which macroscopic raft domains form in GPMVs, with the underlying inference that higher phase separation temperatures suggest more stable domains. This paradigm is based on observations of a specific type of phase separation in GPMVs which occurs near a compositional “critical point” and involves large scale fluctuations at temperatures close the phase transition141. Such “critical fluctuations” are universal to all systems exhibiting critical behavior and suggest scaling laws that allow extrapolation of domain size and stability to living cells135,138. It is important to note that this hypothesis has yet to be formally evaluated; however, if validated, it will provide an important methodological tool for assaying raft properties. For example, it was recently demonstrated that the stability of raft domains in GPMVs is affected by dietary fatty acids. Specifically, exogenously supplied polyunsaturated fatty acids, e.g. the fish oil component ω-3 docosahexaenoic acid, are robustly incorporated into cellular membranes, where they induce extensive remodelling of lipid compositions and biophysical properties, including increasing the stability of raft-like domains67. A study relating these effects to cell behaviour showed that ω-3 docosahexaenoic acid incorporation and concomitant increase in the stability of raft-like domains can repress stem cell properties and motility of breast cancer cells, by interfering with plasma membrane remodelling necessary for the process of epithelial–mesenchymal transition139.
While variations in lipid composition are certainly key drivers of raft behaviour, protein–lipid interactions also play key roles in raft regulation. For example, some proteins, including the HIV glycoprotein gp41140, have cholesterol binding motifs that regulate their membrane distribution (Fig. 4b). Other proteins specifically bind glycosphingolipids141 or sphingomyelin142, potentially mediating their recruitment to membrane domains. Further, a variation on the role of palmitoylation in raft regulation was recently proposed for a post-synaptic density protein 95 (PSD-95). Comprehensive lipidomic analysis of neuronal synapses suggested specific recruitment of raft domains to synaptic sites, which was proposed to be mediated by integration of palmitoylated PSD-95 into the post-synaptic density protein scaffold143. In this formulation, rather than raft domains recruiting palmitoylated proteins, it is the immobilized palmitoylated proteins that recruit saturated lipids and thus nucleate ordered domains at specific cellular sites143 (Fig. 4c). This hypothetical mechanism and its applicability to other cellular contexts remain to be confirmed. However, the evidence that another palmitoylated protein (MPP1) nucleates raft-like environments in erythroid cells144,145 suggests the possibility of a more general mechanism whereby proteins dictate, or at least considerably influence, the localization and stability of organized domains.
Hydrophobic (mis)match
Mammalian membrane lipids can possess hydrocarbon acyl chains from 12 to 24 carbons in length, potentially yielding drastically different hydrophobic tail lengths for individual lipids. To minimize the unfavourable exposure of hydrophobic tails to the aqueous environment, lipids segregate according to their acyl chain length, potentially introducing lateral heterogeneity. In phase-separated model membranes, this thickness mismatch between longer saturated (raft) and shorter unsaturated (non-raft) lipids appears to regulate the size of coexisting domains, with large mismatches giving rise to large domains, and vice versa146. Similarly, TMDs of nearly all eukaryote integral membrane proteins consist of alpha helices with hydrophobic amino-acid side chains, which are buried inside the hydrophobic core of the membrane. Hydrophobic matching between these TMDs and the surrounding membrane lipids minimizes the energetically unfavourable exposure of hydrophobic residues to aqueous environments147 (Fig. 4d). In the case of significant length mismatch between TMDs and their solvating lipids, lateral protein-rich aggregates can potentially be induced148. However, the role of hydrophobic mismatch on membrane domain dynamics in the live cell membrane needs to be unambiguously demonstrated.
Cortical actin cytoskeleton
The cortical actin cytoskeleton is undoubtedly one of the most important factors influencing membrane organization149 and mechanics150. The actin scaffold has been shown to determine molecular diffusion dynamics (e.g. trapped and hop diffusion)151,152 and supramolecular arrangements in the membrane129,153,154. In model membranes, actin can directly stabilize or abrogate large-scale lipid phase separation, depending on the nature of lipid species that are coupled to actin. A holistic model for the interplay between the organization of the cortical actin cytoskeleton and living asymmetric membranes has arisen from studies of the clustering behaviour of GPI-anchored proteins on the outer leaflet of the plasma membrane. It is proposed that such clustering is the result of dynamic self-organization of actin into nanoscopic assemblies termed asters158. These assemblies then, via adaptor proteins (such as ezrin, radixin and moesin), bind the charged lipid phosphatidylserine (PS), which can contain long saturated acyl chains that engage in lipid-mediated interactions with the long acyl chain-containing GPI-anchored proteins located in the opposite leaflet14 (Fig. 4e). Thus, an actin-driven clustering mechanism may be responsible for the formation of ordered domains in live cell membranes, even under conditions (e.g. temperature and/or lipid composition) not normally conducive for phase transitions. A proof-of-principle for this mechanism has been recently demonstrated in vitro159, by showing that dynamically remodelling actomyosin networks can organize an associated synthetic membrane, while these self-organising cortical actin patterns have been shown to associate more ordered membrane environments in the plasma membrane of living cells160. Adding to the chemical principles of lipid–lipid interactions, this actin-driven mechanism of membrane ordering allows to explain the dynamic properties and non-equilibrium distribution of actin–dependent nanoclusters formed by several lipid species, which have been observed in. The molecular machinery generating these actin-based nanoclusters remains as yet unidentified, and further work is necessary to understand how actin-based nanoclusters may give rise to ordered membrane domains161.
Physiological functions of rafts
The most apparent function of raft domains is to segregate specific elements in order to regulate their interactions with other membrane components and hence their activity. Additionally, interactions with raft enriched lipids (cholesterol or glycosphingolipids), or with the distinct biophysical environment of rafts, may change protein conformation and thus bioactivity162 (Fig. 5a). These general modes of regulation may be broadly applied in cellular physiology, and a few examples are described here. However, it should be emphasized that direct mechanistic implications of lipid rafts in cell function and dysfunction remain unclear owing to the inherent difficulties in defining raft composition and properties, and their specific perturbation.
Figure 5. Cellular functions of lipid rafts.
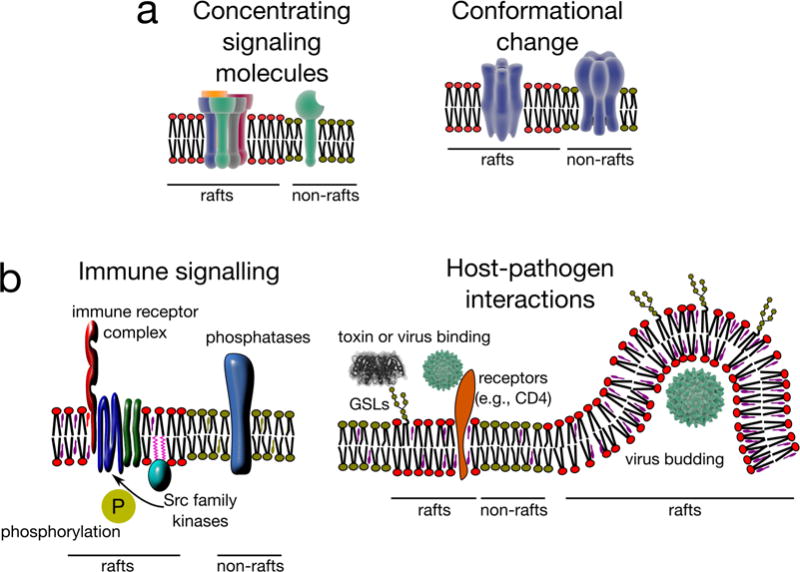
a | Mechanisms by which membrane domains can potentially regulate bioactivity of their associated components. Rafts can concentrate certain molecules resulting in the establishment of functional catalytic platforms. For example, enzyme and substrates can be brought together to increase their encounter probability and thereby trigger reactions (e.g. signal transduction). A related possibility is that distinct physicochemical environments provided by lipid rafts directly impact protein conformation, thereby regulating bioactivity. b | Examples of physiological functions of membrane domains. Kinases of the SRC family are enriched in raft domains owing to their palmitoylation, whereas transmembrane phosphatases are generally excluded. This segregation has been found to be important for immune signalling, where raft associated SRC kinases are involved in regulating the phosphorylation state, and hence the signal transduction activity, of various immune receptors (including the T-cell receptor and the IgE receptor) (left). Many pathogens and their products (such as bacterial toxins) selectively bind membrane rafts owing to the presence of their specific receptors, such as glycoshpingolipids (GSLs; for the cholera toxin) or CD4 (for the human immunodeficiency virus (HIV)) in these domains, thereby gaining access to their host cells. Virus budding is also thought to occur preferentially at raft-like domains. Although the mechanism behind this selective budding is not yet clear, viral proteins such as Gag proteins of HIV are believed to be sensitive to membrane fluidity, and to associate with cholesterol-enriched domains.
Immune signaling
Compartmentalization of cellular signalling in membrane domains may be used to concentrate positive regulatory components (such as kinases163) while excluding negative regulatory elements (such as phosphatases164) (Fig. 5b). The first signalling pathway associated with lipid rafts was IgE-mediated signalling165. Since then, several studies have implicated these domains in a variety of innate and adaptive immune responses166. In these contexts, the key immune receptors, including the IgE receptor (FceRI), T-cell receptor167, and B-cell receptor44, were found in detergent soluble membrane fractions in resting or immature cells, but acquired detergent resistance following receptor activation, suggesting that translocation to membrane rafts is associated with active signalling through these receptors168,169. This notion is supported by co-enrichment in DRMs of the proximal signal transduction machinery downstream of the immune receptors, including lymphocyte-specific protein tyrosine kinase Lck or proto-oncogene tyrosine-protein kinase Fyn163, as well as a signalling adaptor protein LAT43. Further, several other immune-associated proteins are GPI-anchored (suggesting their preferential targeting to rafts) and have been found in DRMs170; these include the receptor for bacterial lipopolysaccharides CD14 and Thy-1 (CD90), which is crucial for T-cell activation171.
Host-pathogen interactions
Recent studies have boosted the interest in lipid rafts as modulator of host– pathogen interactions by disclosing a high content of saturated (sphingo-)lipids and cholesterol in the viral envelopes (for example of HIV)172 or by finding ordered membrane domains in pathogenic microorganisms173. There is now substantial evidence showing that viruses and bacterial products (such as toxins) preferably bind to detergent-resistant and highly ordered plasma membrane regions to penetrate the cell. This could be due to the raft enrichment of receptors, such as glycolipids174 (e.g. for cholera toxin90) or viral receptors175. Further, binding of HIV Gag protein (necessary for viral budding and release from host cells) has been shown to preferentially occur in the membrane domains with high cholesterol content176, suggesting that rafts might be preferential sites for viral budding176(Fig. 5b).
Cancer
A multitude of proteins associated with oncogenic malignancies have been found in DRMs; these include Mucin-1, overexpression of which leads to several cancer forms177, urokinase plasminogen activator receptor, which plays a role in tumour invasion, migration and angiogenesis in breast cancer178, and Ras proteins, which show raft-dependent oncogenic activity in breast cancer179. The localization of pro-oncogenic proteins to raft-like domains, along with the fact that mitogenic signalling is initiated from various cell surface receptors, suggest potential involvement of rafts in cancer development and progression. Consistently, drugs that modulate membrane organization, including the raft-associated alkyl-phospholipids180 edelphosine, miltephosine or perifosine (for instance, by altering raft localisation of proton pumps in the membrane180), have been shown to exhibit anti-cancer activity181.
Cardiovascular diseases
Atherosclerosis is a leading vascular disease, which develops as a result of the uptake of cholesterol accumulated in the artery walls (as oxidized low density lipoproteins (oxLDLs)) by macrophages. This uptake causes a transformation of macrophages into foam cells which then clog the arterial wall leading to strokes, heart attacks and peripheral vascular diseases182. Notably, this transition of macrophages into foam cells appears to be raft-dependent as oxLDL receptors were found to localize to raft domains upon oxLDL stimulation183. In addition, caveolae, formation of which has often been associated with lipid rafts, are also essential for normal cardiac functions, as various cardiac ion channels have been shown to localize to these membrane pits184.
Conclusions and Perspective
Accumulating evidence suggests that cellular membranes are laterally heterogeneous, forming distinct, highly ordered lipid raft domains alongside less organized and more fluid regions. This heterogeneity is potentially important for various cellular functions by regulating the interactions between membrane associated components. However, the mechanisms driving and regulating lateral membrane heterogeneity remain poorly understood. For this reason, the concept of lipid rafts has received an outsized share of both popularity and controversy. At its apex, hundreds of papers per year were published on membrane rafts; at the nadir, many refrained from using the word ‘rafts’ to avoid the inevitable semantic quicksand that it conjured. The major predicament in membrane raft research has been, and continues to be, lack of direct visualization of these domains in unperturbed live cells. However, the remarkable advances in microscopic technology of the last decade now allow direct observation of the spatial (nanometers) and temporal (milliseconds) regimes believed to be relevant for raft domains in the living cells. These advances suggest that direct of detection of these elusive domains in cell membranes, while remaining challenging, may be within reach185. Direct imaging of phase separation in isolated plasma membranes, such as GPMVs37,38, has already provided evidence that the isolated plasma membrane bilayer is capable of generating co-existing liquid ordered and disordered domains. Moreover, domains remarkably resembling those ordered and disordered phase separated domains in GPMVs have been directly visualized in subcellular organelles of yeast186, suggesting that investigation of internal membranes may also be a fruitful direction.
Much of the controversy about the properties (size, lifetime, abundance) of lipid rafts stems from attempts to generalize the organization of a number of different membrane components (incl. glycolipids, sphingomyelin, cholesterol, GPI-anchored proteins, and minimal palmitoylated motifs) via a common raft paradigm. First, it is important to note that a very specific set of physical and compositional features should not be expected from lipid rafts. Living membranes are extremely complex and varied, and thus their organization will inherently be context-dependent, and potentially involve many different types of coexisting domains. Such varied assemblies may have distinct organizational principles and cellular functions, which may only be apparent at specific spatial and temporal scales. Moreover, it is important to consider that most molecules that typically associate with rafts are not simply domain probes, but also possess distinct bioactivities that may impact domain organization and dynamics. Further, these bioactivities may be affected by the specifics of an experiment, e.g. cell type or cell cylce. Altogether, to obtain reproducible results regarding raft formation and their biophysical properties it may be necessary to introduce fully synthetic probes (instead of semi-native labels) that exhibit validated affinities for ordered membrane domains84, and thus allow to careful correlations between ordered domain affinity and other experimental readouts22. Another approach that would minimize the experimental differences is the application of label-free detection of domains72.
Ultimately, the controversies about the organization and dynamics of membrane domains will be resolved by direct observation of well-validated probes, with high spatial and temporal resolution, over extended time- and length-scales. Such data could be complemented by detailed lipidomic and proteomic analysis of nanometric regions of the cell surface70. The next step will be to integrate these observations into the framework of cellular dynamics, linking membrane heterogeneity to cell biological processes. For this, it will be necessary to concomitantly observe the organization, dynamics, and bioactivity of specific raft components to dissect the key principles of how domain localization modulates molecular function. Clearly, such advances will require parallel application and development of a variety of different techniques, suggesting that this field has an exciting and interdisciplinary future.
Supplementary Material
Online summary.
Cellular membranes are lateral heterogeneous, consisting of transient and dynamic domains with varying properties, prominently including ordered lipid-driven domains referred to as lipid (or membrane) rafts.
Membrane domains can be induced and regulated by a variety of interactions, including specific lipid–lipid and lipid–protein interactions, bulk membrane properties, and interactions between membrane components and the underlying cytoskeleton.
Advanced microscopy and biochemistry techniques facilitate the study of membrane domains, however they still elude direct in vivo visualization. The multiplicity of possible organizational states and their context-dependent nature most likely account for experimental inconsistencies.
Membrane rafts potentially play crucial physiological roles across cell types, spanning from immune cells to cancer cells.
Membrane domains are conserved throughout the domains of life, supporting their important functions in biological systems.
Acknowledgments
CE and ES are supported by Wolfson Foundation, the Medical Research Council (MRC, grant number MC_UU_12010/unit programmes G0902418 and MC_UU_12025), MRC/BBSRC/ESPRC (grant number MR/K01577X/1), and the Wellcome Trust (grant ref 104924/14/Z/14). ES is supported by EMBO Long Term (ALTF 636–2013) and Marie Curie Intra-European Fellowships (MEMBRANE DYNAMICS). SM is supported by a JC Bose fellowship from the Department of Science and Technology and a Margadarshi fellowship (DBT-Wellcome Trust Alliance grant ref IA/M/15/1/502018). IL is supported by the Cancer Prevention and Research Institute of Texas (R1215) and the National Institutes of Health (grant 1R01GM114282).
Glossary
- liquid–liquid phase separation
coexistence of two phases with distinct compositions and biophysical properties. The components of both phases can diffuse and rearrange rapidly
- sphingolipid
a class of lipids based on a long chain sphingosine base coupled to a fatty acid chain and often a large polar headgroup
- glycophosphatidylinositol-anchored proteins
cell surface proteins that are post-translationally modified to carry a glycophosphatidylinositol moiety as an anchor to the membrane
- cholera toxin
proteinaceous toxin secreted by Vibrio cholerea that binds glycolipids on the cell surface and is responsible for cholera infection
- single particle tracking (SPT)
a single molecule technique where the motion of individual molecules is tracked with high temporal resolution over relatively long time scales (seconds); these tracks can be used to determine the molecules’ diffusion properties.
- confined diffusion
also known as trapped diffusion; a diffusion mode where the motion of the molecule is transiently arrested by molecular obstacles such as immobile clusters.
- hop diffusion
a diffusion mode where molecules diffuse freely on the membrane except when encountering a barrier, such as structures associated with actin filaments
- interferometric scattering (iSCAT) microscopy
a microscopy technique wherein interference from coherent light scattering in the focal plane and of the microscope cover glass is used to enhance contrast
- fluorescence correlation spectroscopy
a single molecule-based technique wherein fluorescence intensity fluctuations from a microscopic observation spot are used to obtain information about molecular diffusion
- Förster resonance energy transfer (FRET)
a fluorescence spectroscopy and imaging technique based on the distance-dependent transfer of the excited state energy of a donor fluorescent molecule to an acceptor fluorescent molecule; efficient and widely used to measure intermolecular distances in the range of 1–10 nm.
- amphiphilic molecules
molecules showing both hydrophilic and hydrophobic character, such as lipids with hydrophobic acyl chains and hydrophilic headgroups
- Raman spectroscopy
a spectroscopy technique where vibrational energy of the molecules is used as fingerprints of the molecules
- ganglioside lipid
a class of glycosphingolipid with sialic acid moieties on the head group
- ceramides
a class of lipids composed of sphingosine and a fatty acid
- coarse-grained simulations
simulations that rely on simplified representations of the simulated components
- hydrogen-bonding
non-covalent chemical bonds formed between a hydrogen covalently bound to a electronegative atom (as in the -NH group of sphingolipids) and another electronegative atom (such as the oxygen in the -OH group of cholesterol)
- epithelial–mesenchymal transition
a developmental transcriptional program that imparts mesenchymal characteristics (e.g. motility) to epithelial cells
- viral envelope
the lipid membrane that covers the virus capsid protein and is derived from the host cell plasma membrane
- caveolae
specialized invaginations in the plasma membrane enriched in caveolin, sphingolipids, and cholesterol
Biographies
Erdinc Sezgin has been an EMBO and a Marie Skłodowska-Curie fellow in University of Oxford since 2014. He carried out his PhD work in the group of Petra Schwille at the Technical University of Dresden and a short postdoctoral period in Kai Simons lab at Max Planck Institute of Cell Biology and Genetics in Dresden, Germany. His research is focused on the role of membrane heterogeneity in immune system.
Ilya Levental received his PhD from the University of Pennsylvania under the guidance of Dr Paul Janmey. His postdoctoral research on the mechanisms of protein recruitment to membrane domains was carried out during a Humboldt Fellowship in the laboratory of Dr Kai Simons at the MPI for Molecular Cell Biology and Genetics (MPI-CBG) in Dresden, Germany. In 2012, he became a group leader and CPRIT scholar for cancer research at the McGovern Medical School of the University of Texas Health Science Center at Houston. His research continues to focus on the physiological consequences of membrane composition and organization, with specific focus on dietary lipids.
Satyajit Mayor studied chemistry at the Indian Institute of Technology Bombay and was awarded his Ph.D. from The Rockefeller University. He worked as a post-doctoral fellow at Columbia University. He is currently Senior Professor and is Centre Director of the National Centre for Biological Sciences, Bangalore, and the Director of the Institute for Stem Cell Biology and Regenerative Medicine at Bangalore.
Christian Eggeling holds a diploma and PhD in Physics from the Universities of Hamburg and Göttingen. Between 2000–2003, he worked at Evotec, Hamburg. End of 2003, he joined Prof. Stefan Hell’s lab at the Max-Planck-Institute of Biophysical Chemistry in Gottingen, Germany. Since 2012, he is a principal investigator (and professor since 2014) in the MRC Human Immunology Unit and the scientific director of the Wolfson Imaging Centre at the Weatherall Institute of Molecular Medicine at the University of Oxford.
Contributor Information
Erdinc Sezgin, MRC Human Immunology Unit, OX39DS, University of Oxford, UK.
Ilya Levental, Department of Integrative Biology and Pharmacology, University of Texas Health Science Center, United States.
Satyajit Mayor, National Centre for Biological Sciences, Tata Institute for Fundamental Research, Bellary Road, GKVK, 560065 Bangalore, India, Tel: 0091 80 2366 6260.
Christian Eggeling, MRC Human Immunology Unit, OX39DS, University of Oxford, UK, Tel: 0044 1865 222167.
References
- 1.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–&. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Fischman DA, Steck TL. Selective solubilization of proteins and phospholipids from red blood cell membranes by nonionic detergents. Journal of Supramolecular Structure. 1973;1:233–248. doi: 10.1002/jss.400010308. [DOI] [PubMed] [Google Scholar]
- 3.Bagatolli L, Mouritsen O. Is the fluid mosaic (and the accompanying raft hypothesis) a suitable model to describe fundamental features of biological membranes? What may be missing? Frontiers in Plant Science. 2013;4 doi: 10.3389/fpls.2013.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed SN, Brown DA, London E. On the origin of sphingolipid/cholesterol-rich detergent-insoluble cell membranes: Physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble, liquid-ordered lipid phase in model membranes. Biochemistry. 1997;36:10944–10953. doi: 10.1021/bi971167g. [DOI] [PubMed] [Google Scholar]
- 5.Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 6.Vanmeer G, Stelzer EHK, Wijnaendtsvanresandt RW, Simons K. Sorting of sphingolipids in epithelial (Madin-Darby canine kidney) cells. Journal of Cell Biology. 1987;105:1623–1635. doi: 10.1083/jcb.105.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varma R, Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- 8.Pralle A, Keller P, Florin EL, Simons K, Horber JK. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. The Journal of cell biology. 2000;148:997–1008. doi: 10.1083/jcb.148.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedrichson T, Kurzchalia TV. Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature. 1998;394:802–805. doi: 10.1038/29570. [DOI] [PubMed] [Google Scholar]
- 10.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 11.Simons K, Vaz WL. Model systems, lipid rafts, and cell membranes. Annual review of biophysics and biomolecular structure. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 12.Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Kiessling V, Wan C, Tamm LK. Domain coupling in asymmetric lipid bilayers. Biochim Biophys Acta. 2009;1788:64–71. doi: 10.1016/j.bbamem.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghupathy R, et al. Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell. 2015;161:581–594. doi: 10.1016/j.cell.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 16.Klotzsch E, Schuetz GJ. A critical survey of methods to detect plasma membrane rafts. Philosophical Transactions of the Royal Society B-Biological Sciences. 2013;368:20120033. doi: 10.1098/rstb.2012.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanada K, Nishijima M, Akamatsu Y, Pagano RE. Both sphingolipids and cholesterol participate in the detergent insolubility of alkaline phosphatase, a glycosylphosphatidylinositol-anchored protein, in mammalian membranes. J Biol Chem. 1995;270:6254–6260. doi: 10.1074/jbc.270.11.6254. [DOI] [PubMed] [Google Scholar]
- 18.Schroeder R, London E, Brown D. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc Natl Acad Sci U S A. 1994;91:12130–12134. doi: 10.1073/pnas.91.25.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K. Resistance of cell membranes to different detergents. Proc Natl Acad Sci U S A. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayor S, Maxfield FR. Insolubility and redistribution of GPI-anchored proteins at the cell surface after detergent treatment. Mol Biol Cell. 1995;6:929–944. doi: 10.1091/mbc.6.7.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levental I, et al. Cholesterol-dependent phase separation in cell-derived giant plasma-membrane vesicles. Biochem J. 2009;424:163–167. doi: 10.1042/BJ20091283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komura N, et al. Raft-based interactions of gangliosides with a GPI-anchored receptor. Nat Chem Biol. 2016;12:402–410. doi: 10.1038/nchembio.2059. [DOI] [PubMed] [Google Scholar]
- 23.Lichtenberg D, Goni FM, Heerklotz H. Detergent-resistant membranes should not be identified with membrane rafts. Trends in biochemical sciences. 2005;30:430–436. doi: 10.1016/j.tibs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Sezgin E, Schwille P. Model membrane platforms to study protein-membrane interactions. Molecular Membrane Biology. 2012;29:144–154. doi: 10.3109/09687688.2012.700490. [DOI] [PubMed] [Google Scholar]
- 25.Ipsen JH, Karlstrom G, Mouritsen OG, Wennerstrom H, Zuckermann MJ. Phase equilibria in the phosphatidylcholine-cholesterol system. Biochim Biophys Acta. 1987;905:162–172. doi: 10.1016/0005-2736(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 26.Veatch SL, Keller SL. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaiser HJ, et al. Order of lipid phases in model and plasma membranes. Proc Natl Acad Sci U S A. 2009;106:16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McConnell HM, Tamm LK, Weis RM. Periodic structures in lipid monolayer phase transitions. Proceedings of the National Academy of Sciences of the United States of America-Physical Sciences. 1984;81:3249–3253. doi: 10.1073/pnas.81.10.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamm LK, McConnell HM. Supported phospholipid-bilayers. Biophysical Journal. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feigenson GW, Buboltz JT. Ternary phase diagram of dipalmitoyl-PC/dilauroyl-PC/cholesterol: nanoscopic domain formation driven by cholesterol. Biophys J. 2001;80:2775–2788. doi: 10.1016/S0006-3495(01)76245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korlach J, Schwille P, Webb WW, Feigenson GW. Characterization of lipid bilayer phases by confocal microscopy and fluorescence correlation spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8461–8466. doi: 10.1073/pnas.96.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagatolli LA, Sanchez SA, Hazlett T, Gratton E. Giant vesicles, Laurdan, and two-photon fluorescence microscopy: evidence of lipid lateral separation in bilayers. Methods Enzymology. 2003;360:481–500. doi: 10.1016/s0076-6879(03)60124-2. [DOI] [PubMed] [Google Scholar]
- 33.Kahya N, Brown DA, Schwille P. Raft partitioning and dynamic behavior of human placental alkaline phosphatase in giant unilamellar vesicles. Biochemistry. 2005;44:7479–7489. doi: 10.1021/bi047429d. [DOI] [PubMed] [Google Scholar]
- 34.Stachowiak JC, et al. Unilamellar vesicle formation and encapsulation by microfluidic jetting. Proc Natl Acad Sci U S A. 2008;105:4697–4702. doi: 10.1073/pnas.0710875105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupuy AD, Engelman DM. Protein area occupancy at the center of the red blood cell membrane. Proc Natl Acad Sci U S A. 2008;105:2848–2852. doi: 10.1073/pnas.0712379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sezgin E, et al. Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochimica Et Biophysica Acta-Biomembranes. 2012;1818:1777–1784. doi: 10.1016/j.bbamem.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Sezgin E, et al. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nature Protocols. 2012;7:1042–1051. doi: 10.1038/nprot.2012.059. [DOI] [PubMed] [Google Scholar]
- 38.Baumgart T, et al. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci U S A. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fridriksson EK, et al. Quantitative analysis of phospholipids in functionally important membrane domains from RBL-2H3 mast cells using tandem high-resolution mass spectrometry. Biochemistry. 1999;38:8056–8063. doi: 10.1021/bi9828324. [DOI] [PubMed] [Google Scholar]
- 40.Scott RE, Perkins RG, Zschunke MA, Hoerl BJ, Maercklein PB. Plasma membrane vesiculation in 3T3 and SV3T3 cells. I. Morphological and biochemical characterization. J Cell Sci. 1979;35:229–243. doi: 10.1242/jcs.35.1.229. [DOI] [PubMed] [Google Scholar]
- 41.Keller H, Lorizate M, Schwille P. PI(4,5)P-2 Degradation Promotes the Formation of Cytoskeleton-Free Model Membrane Systems. Chemphyschem. 2009;10:2805–2812. doi: 10.1002/cphc.200900598. [DOI] [PubMed] [Google Scholar]
- 42.Levental KR, Levental I. Giant plasma membrane vesicles: models for understanding membrane organization. Current topics in membranes. 2015;75:25–57. doi: 10.1016/bs.ctm.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 43.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta N, DeFranco AL. Visualizing lipid raft dynamics and early signaling events during antigen receptor-mediated B-lymphocyte activation. Mol Biol Cell. 2003;14:432–444. doi: 10.1091/mbc.02-05-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sezgin E, Schwille P. Fluorescence Techniques to Study Lipid Dynamics. Cold Spring Harbor Perspectives in Biology. 2011;3:a009803. doi: 10.1101/cshperspect.a009803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eggeling C. Super-resolution optical microscopy of lipid plasma membrane dynamics. Essays in biochemistry. 2015;57:69–80. doi: 10.1042/bse0570069. [DOI] [PubMed] [Google Scholar]
- 47.Owen DM, Williamson DJ, Magenau A, Gaus K. Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nature Communications. 2012;3 doi: 10.1038/ncomms2273. [DOI] [PubMed] [Google Scholar]
- 48.Sengupta P, et al. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nature methods. 2011;8:969–975. doi: 10.1038/nmeth.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Zanten TS, et al. Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc Natl Acad Sci U S A. 2009;106:18557–18562. doi: 10.1073/pnas.0905217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saka SK, et al. Multi-protein assemblies underlie the mesoscale organization of the plasma membrane. Nature Communications. 2014;5 doi: 10.1038/ncomms5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki KG. Single-Molecule Imaging of Signal Transduction via GPI-Anchored Receptors. Methods in molecular biology (Clifton, NJ) 2016;1376:229–238. doi: 10.1007/978-1-4939-3170-5_19. [DOI] [PubMed] [Google Scholar]
- 52.Moertelmaier M, Brameshuber M, Linimeier M, Schütz GJ, Stockinger H. Thinning out clusters while conserving stoichiometry of labeling. Applied Physics Letters. 2005;87:263903. [Google Scholar]
- 53.Kusumi A, et al. Annual review of biophysics and biomolecular structure Vol.34 Annual Review of Biophysics. 2005:351–U354. [Google Scholar]
- 54.Ortega-Arroyo J, Kukura P. Interferometric scattering microscopy (iSCAT): new frontiers in ultrafast and ultrasensitive optical microscopy. Physical chemistry chemical physics: PCCP. 2012;14:15625–15636. doi: 10.1039/c2cp41013c. [DOI] [PubMed] [Google Scholar]
- 55.Wu HM, Lin YH, Yen TC, Hsieh CL. Nanoscopic substructures of raft-mimetic liquid-ordered membrane domains revealed by high-speed single-particle tracking. Scientific reports. 2016;6:20542. doi: 10.1038/srep20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spillane KM, et al. High-speed single-particle tracking of GM1 in model membranes reveals anomalous diffusion due to interleaflet coupling and molecular pinning. Nano letters. 2014;14:5390–5397. doi: 10.1021/nl502536u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wawrezinieck L, Rigneault H, Marguet D, Lenne PF. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys J. 2005;89:4029–4042. doi: 10.1529/biophysj.105.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eggeling C, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 59.Honigmann A, et al. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nature Communications. 2014;5:5412–5412. doi: 10.1038/ncomms6412. [DOI] [PubMed] [Google Scholar]
- 60.Saha S, Raghupathy R, Mayor S. Homo-FRET imaging highlights the nanoscale organization of cell surface molecules. Methods in molecular biology (Clifton, NJ) 2015;1251:151–173. doi: 10.1007/978-1-4939-2080-8_9. [DOI] [PubMed] [Google Scholar]
- 61.Sharma P, et al. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 62.Pathak P, London E. The Effect of Membrane Lipid Composition on the Formation of Lipid Ultrananodomains. Biophys J. 2015;109:1630–1638. doi: 10.1016/j.bpj.2015.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Engel S, et al. FLIM-FRET and FRAP reveal association of influenza virus haemagglutinin with membrane rafts. Biochem J. 2010;425:567–573. doi: 10.1042/BJ20091388. [DOI] [PubMed] [Google Scholar]
- 64.Heberle FA, Wu J, Goh SL, Petruzielo RS, Feigenson GW. Comparison of three ternary lipid bilayer mixtures: FRET and ESR reveal nanodomains. Biophys J. 2010;99:3309–3318. doi: 10.1016/j.bpj.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barrera NP, Zhou M, Robinson CV. The role of lipids in defining membrane protein interactions: insights from mass spectrometry. Trends in cell biology. 2013;23:1–8. doi: 10.1016/j.tcb.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 66.Ogiso H, Taniguchi M, Okazaki T. Analysis of lipid-composition changes in plasma membrane microdomains. J Lipid Res. 2015;56:1594–1605. doi: 10.1194/jlr.M059972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levental KR, et al. Polyunsaturated lipids regulate membrane domain stability by tuning membrane order. Biophys J. 2016;110(8):1800–1810. doi: 10.1016/j.bpj.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gerl MJ, et al. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. The Journal of cell biology. 2012;196:213–221. doi: 10.1083/jcb.201108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lozano MM, Hovis JS, Moss FR, 3rd, Boxer SG. Dynamic Reorganization and Correlation among Lipid Raft Components. J Am Chem Soc. 2016;138:9996–10001. doi: 10.1021/jacs.6b05540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lozano MM, et al. Colocalization of the ganglioside G(M1) and cholesterol detected by secondary ion mass spectrometry. J Am Chem Soc. 2013;135:5620–5630. doi: 10.1021/ja310831m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frisz JF, et al. Sphingolipid domains in the plasma membranes of fibroblasts are not enriched with cholesterol. Journal of Biological Chemistry. 2013;288:16855–16861. doi: 10.1074/jbc.M113.473207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Wit G, Danial JS, Kukura P, Wallace MI. Dynamic label-free imaging of lipid nanodomains. Proc Natl Acad Sci U S A. 2015;112:12299–12303. doi: 10.1073/pnas.1508483112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ando J, et al. Sphingomyelin distribution in lipid rafts of artificial monolayer membranes visualized by Raman microscopy. Proc Natl Acad Sci U S A. 2015;112:4558–4563. doi: 10.1073/pnas.1418088112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pencer J, Mills TT, Kucerka N, Nieh MP, Katsaras J. Small-angle neutron scattering to detect rafts and lipid domains. Methods in molecular biology (Clifton, NJ) 2007;398:231–244. doi: 10.1007/978-1-59745-513-8_16. [DOI] [PubMed] [Google Scholar]
- 75.Heberle FA, et al. Hybrid and nonhybrid lipids exert common effects on membrane raft size and morphology. J Am Chem Soc. 2013;135:14932–14935. doi: 10.1021/ja407624c. [DOI] [PubMed] [Google Scholar]
- 76.Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baumgart T, Hunt G, Farkas ER, Webb WW, Feigenson GW. Fluorescence probe partitioning between L-o/L-d phases in lipid membranes. Biochimica Et Biophysica Acta-Biomembranes. 2007;1768:2182–2194. doi: 10.1016/j.bbamem.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klymchenko AS, Kreder R. Fluorescent probes for lipid rafts: from model membranes to living cells. Chemistry & Biology. 2014;21:97–113. doi: 10.1016/j.chembiol.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 79.Juhasz J, Davis JH, Sharom FJ. Fluorescent probe partitioning in giant unilamellar vesicles of ‘lipid raft’ mixtures. Biochemical Journal. 2010;430:415–423. doi: 10.1042/BJ20100516. [DOI] [PubMed] [Google Scholar]
- 80.Pourmousa M, et al. Dehydroergosterol as an Analogue for Cholesterol: Why It Mimics Cholesterol So Well-or Does It? Journal of Physical Chemistry B. 2014;118:7345–7357. doi: 10.1021/jp406883k. [DOI] [PubMed] [Google Scholar]
- 81.Robalo JR, Martins do Canto AMT, Palace Carvalho AJ, Prates Ramalho JP, Loura LMS. Behavior of Fluorescent Cholesterol Analogues Dehydroergosterol and Cholestatrienol in Lipid Bilayers: A Molecular Dynamics Study. Journal of Physical Chemistry B. 2013;117:5806–5819. doi: 10.1021/jp312026u. [DOI] [PubMed] [Google Scholar]
- 82.Sezgin E, et al. A comparative study on fluorescent cholesterol analogs as versatile cellular reporters. J Lipid Res. 2016;57:299–309. doi: 10.1194/jlr.M065326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crane JM, Tamm LK. Role of cholesterol in the formation and nature of lipid rafts in planar and spherical model membranes. Biophys J. 2004;86:2965–2979. doi: 10.1016/S0006-3495(04)74347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Momin N, et al. Designing lipids for selective partitioning into liquid ordered membrane domains. Soft Matter. 2015;11:3241–3250. doi: 10.1039/c4sm02856b. [DOI] [PubMed] [Google Scholar]
- 85.Lopes SC, Goormaghtigh E, Cabral BJ, Castanho MA. Filipin orientation revealed by linear dichroism. Implication for a model of action. J Am Chem Soc. 2004;126:5396–5402. doi: 10.1021/ja031782+. [DOI] [PubMed] [Google Scholar]
- 86.Palmer M. The family of thiol-activated, cholesterol-binding cytolysins. Toxicon: official journal of the International Society on Toxinology. 2001;39:1681–1689. doi: 10.1016/s0041-0101(01)00155-6. [DOI] [PubMed] [Google Scholar]
- 87.Skocaj M, et al. Tracking cholesterol/sphingomyelin-rich membrane domains with the ostreolysin A-mCherry protein. PLoS One. 2014;9:e92783. doi: 10.1371/journal.pone.0092783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamaji A, et al. Lysenin, a novel sphingomyelin-specific binding protein. J Biol Chem. 1998;273:5300–5306. doi: 10.1074/jbc.273.9.5300. [DOI] [PubMed] [Google Scholar]
- 89.Bhat HB, et al. Binding of a pleurotolysin ortholog from Pleurotus eryngii to sphingomyelin and cholesterol-rich membrane domains. J Lipid Res. 2013;54:2933–2943. doi: 10.1194/jlr.D041731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harder T, Scheiffele P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. Journal of Cell Biology. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dietrich C, et al. Lipid rafts reconstituted in model membranes. Biophysical Journal. 2001;80:1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sezgin E, et al. Adaptive lipid packing and bioactivity in membrane domains. PLoS One. 2015;10:e0123930. doi: 10.1371/journal.pone.0123930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gray EM, Diaz-Vazquez G, Veatch SL. Growth Conditions and Cell Cycle Phase Modulate Phase Transition Temperatures in RBL-2H3 Derived Plasma Membrane Vesicles. PloS one. 2015;10:e0137741. doi: 10.1371/journal.pone.0137741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sezgin E, Sadowski T, Simons K. Measuring Lipid Packing of Model and Cellular Membranes with Environment Sensitive Probes. Langmuir. 2014;30:8160–8166. doi: 10.1021/la501226v. [DOI] [PubMed] [Google Scholar]
- 95.Sezgin E, Waithe D, Bernardino de la Serna J, Eggeling C. Spectral imaging to measure heterogeneity in membrane lipid packing. Chemphyschem. 2015;16:1387–1394. doi: 10.1002/cphc.201402794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parasassi T, Krasnowska EK, Bagatolli L, Gratton E. LAURDAN and PRODAN as polarity-sensitive fluorescent membrane probes. Journal of Fluorescence. 1998;8:365–373. [Google Scholar]
- 97.Parasassi T, De Stasio G, Ravagnan G, Rusch RM, Gratton E. Quantitation of lipid phases in phospholipid vesicles by the generalized polarization of Laurdan fluorescence. Biophysical Journal. 1991;60:179–189. doi: 10.1016/S0006-3495(91)82041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanchez SA, Tricerri MA, Gratton E. Laurdan generalized polarization fluctuations measures membrane packing micro-heterogeneity in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7314–7319. doi: 10.1073/pnas.1118288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Golfetto O, Hinde E, Gratton E. Laurdan fluorescence lifetime discriminates cholesterol content from changes in fluidity in living cell membranes. Biophysical Journal. 2013;104:1238–1247. doi: 10.1016/j.bpj.2012.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kreder R, et al. Solvatochromic Nile Red probes with FRET quencher reveal lipid order heterogeneity in living and apoptotic cells. ACS Chem Biol. 2015;10:1435–1442. doi: 10.1021/cb500922m. [DOI] [PubMed] [Google Scholar]
- 101.Mahammad S, Parmryd I. Cholesterol depletion using methyl-beta-cyclodextrin. Methods in molecular biology (Clifton, NJ) 2015;1232:91–102. doi: 10.1007/978-1-4939-1752-5_8. [DOI] [PubMed] [Google Scholar]
- 102.Pottosin II, Valencia-Cruz G, Bonales-Alatorre E, Shabala SN, Dobrovinskaya OR. Methyl-beta-cyclodextrin reversibly alters the gating of lipid rafts-associated Kv1.3 channels in Jurkat T lymphocytes. Pflugers Arch. 2007;454:235–244. doi: 10.1007/s00424-007-0208-4. [DOI] [PubMed] [Google Scholar]
- 103.Mahammad S, Dinic J, Adler J, Parmryd I. Limited cholesterol depletion causes aggregation of plasma membrane lipid rafts inducing T cell activation. Biochim Biophys Acta. 2010;1801:625–634. doi: 10.1016/j.bbalip.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 104.Sanchez SA, Gunther G, Tricerri MA, Gratton E. Methyl-beta-cyclodextrins preferentially remove cholesterol from the liquid disordered phase in giant unilamellar vesicles. The Journal of membrane biology. 2011;241:1–10. doi: 10.1007/s00232-011-9348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hillyard DZ, et al. Statins inhibit NK cell cytotoxicity by membrane raft depletion rather than inhibition of isoprenylation. Atherosclerosis. 2007;191:319–325. doi: 10.1016/j.atherosclerosis.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 106.Amin D, et al. RPR 107393, a potent squalene synthase inhibitor and orally effective cholesterol-lowering agent: comparison with inhibitors of HMG-CoA reductase. The Journal of pharmacology and experimental therapeutics. 1997;281:746–752. [PubMed] [Google Scholar]
- 107.Ahn KW, Sampson NS. Cholesterol oxidase senses subtle changes in lipid bilayer structure. Biochemistry. 2004;43:827–836. doi: 10.1021/bi035697q. [DOI] [PubMed] [Google Scholar]
- 108.Merrill AH, Jr, van Echten G, Wang E, Sandhoff K. Fumonisin B1 inhibits sphingosine (sphinganine) Nacyltransferase and de novo sphingolipid biosynthesis in cultured neurons in situ. J Biol Chem. 1993;268:27299–27306. [PubMed] [Google Scholar]
- 109.Zhao Y, et al. ABCB4 exports phosphatidylcholine in a sphingomyelin-dependent manner. J Lipid Res. 2015;56:644–652. doi: 10.1194/jlr.M056622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miller H, et al. Lipid raft-dependent plasma membrane repair interferes with the activation of B lymphocytes. The Journal of cell biology. 2015;211:1193–1205. doi: 10.1083/jcb.201505030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eggeling C, Honigmann A. Closing the gap: The approach of optical and computational microscopy to uncover biomembrane organization. Biochim Biophys Acta. 2016;1858:2558–2568. doi: 10.1016/j.bbamem.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 112.Dror RO, Dirks RM, Grossman JP, Xu H, Shaw DE. Biomolecular simulation: a computational microscope for molecular biology. Annual review of biophysics. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 113.Lane TJ, Shukla D, Beauchamp KA, Pande VS. To milliseconds and beyond: challenges in the simulation of protein folding. Current opinion in structural biology. 2013;23:58–65. doi: 10.1016/j.sbi.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Saunders MG, Voth GA. Coarse-graining methods for computational biology. Annual review of biophysics. 2013;42:73–93. doi: 10.1146/annurev-biophys-083012-130348. [DOI] [PubMed] [Google Scholar]
- 115.Vattulainen I, Rog T. Lipid simulations: a perspective on lipids in action. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stansfeld PJ, Sansom MS. Molecular simulation approaches to membrane proteins. Structure (London, England: 1993) 2011;19:1562–1572. doi: 10.1016/j.str.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 117.Ingolfsson HI, et al. Lipid organization of the plasma membrane. J Am Chem Soc. 2014;136:14554–14559. doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- 118.Niemela PS, Ollila S, Hyvonen MT, Karttunen M, Vattulainen I. Assessing the nature of lipid raft membranes. PLoS computational biology. 2007;3:e34. doi: 10.1371/journal.pcbi.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Barua D, Goldstein B. A mechanistic model of early FcepsilonRI signaling: lipid rafts and the question of protection from dephosphorylation. PLoS One. 2012;7:e51669. doi: 10.1371/journal.pone.0051669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Levental I, Grzybek M, Simons K. Raft domains of variable properties and compositions in plasma membrane vesicles. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11411–11416. doi: 10.1073/pnas.1105996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schutz GJ, Kada G, Pastushenko VP, Schindler H. Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. The EMBO journal. 2000;19:892–901. doi: 10.1093/emboj/19.5.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang TY, Silvius JR. Sphingolipid partitioning into ordered domains in cholesterol-free and cholesterol-containing lipid bilayers. Biophys J. 2003;84:367–378. doi: 10.1016/S0006-3495(03)74857-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sodt AJ, Pastor RW, Lyman E. Hexagonal Substructure and Hydrogen Bonding in Liquid-Ordered Phases Containing Palmitoyl Sphingomyelin. Biophys J. 2015;109:948–955. doi: 10.1016/j.bpj.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ramstedt B, Slotte JP. Interaction of cholesterol with sphingomyelins and acyl-chain-matched phosphatidylcholines: a comparative study of the effect of the chain length. Biophys J. 1999;76:908–915. doi: 10.1016/S0006-3495(99)77254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sodt AJ, Sandar ML, Gawrisch K, Pastor RW, Lyman E. The molecular structure of the liquid-ordered phase of lipid bilayers. J Am Chem Soc. 2014;136:725–732. doi: 10.1021/ja4105667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Levental I, Grzybek M, Simons K. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry. 2010;49:6305–6316. doi: 10.1021/bi100882y. [DOI] [PubMed] [Google Scholar]
- 127.Ozhan G, et al. Lypd6 Enhances Wnt/beta-Catenin Signaling by Promoting Lrp6 Phosphorylation in Raft Plasma Membrane Domains. Developmental Cell. 2013;26:331–345. doi: 10.1016/j.devcel.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 128.Brameshuber M, et al. Imaging of Mobile Long-lived Nanoplatforms in the Live Cell Plasma Membrane. Journal of Biological Chemistry. 2010;285:41765–41771. doi: 10.1074/jbc.M110.182121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Goswami D, et al. Nanoclusters of GPI-Anchored Proteins Are Formed by Cortical Actin-Driven Activity. Cell. 2008;135:1085–1097. doi: 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sevcsik E, et al. GPI-anchored proteins do not reside in ordered domains in the live cell plasma membrane. Nature Communications. 2015;6:6969. doi: 10.1038/ncomms7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shi D, et al. Smoothened oligomerization/higher order clustering in lipid rafts is essential for high Hedgehog activity transduction. J Biol Chem. 2013;288:12605–12614. doi: 10.1074/jbc.M112.399477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shah A, et al. RaftProt: mammalian lipid raft proteome database. Nucleic acids research. 2015;43:D335–338. doi: 10.1093/nar/gku1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lorent JH, Levental I. Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem Phys Lipids. 2015;192:23–32. doi: 10.1016/j.chemphyslip.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 134.Diaz-Rohrer BB, Levental KR, Simons K, Levental I. Membrane raft association is a determinant of plasma membrane localization. Proc Natl Acad Sci U S A. 2014;111:8500–8505. doi: 10.1073/pnas.1404582111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Veatch SL, et al. Critical fluctuations in plasma membrane vesicles. Acs Chemical Biology. 2008;3:287–293. doi: 10.1021/cb800012x. [DOI] [PubMed] [Google Scholar]
- 136.Lingwood D, Ries J, Schwille P, Simons K. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10005–10010. doi: 10.1073/pnas.0804374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhou Y, et al. Bile acids modulate signaling by functional perturbation of plasma membrane domains. J Biol Chem. 2013;288:35660–35670. doi: 10.1074/jbc.M113.519116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Levental I, Veatch S. The continuing mystery of lipid rafts. Journal of molecular biology. 2016 doi: 10.1016/j.jmb.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tisza MJ, et al. Motility and stem cell properties induced by the epithelial-mesenchymal transition require destabilization of lipid rafts. Oncotarget. 2016 doi: 10.18632/oncotarget.9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schwarzer R, et al. The cholesterol-binding motif of the HIV-1 glycoprotein gp41 regulates lateral sorting and oligomerization. Cellular microbiology. 2014;16:1565–1581. doi: 10.1111/cmi.12314. [DOI] [PubMed] [Google Scholar]
- 141.Coskun U, Grzybek M, Drechsel D, Simons K. Regulation of human EGF receptor by lipids. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1105666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Contreras FX, et al. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature. 2012;481:525–529. doi: 10.1038/nature10742. [DOI] [PubMed] [Google Scholar]
- 143.Tulodziecka K, et al. Remodeling of the postsynaptic plasma membrane during neural development. Mol Biol Cell. 2016;27:3480–3489. doi: 10.1091/mbc.E16-06-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lach A, et al. Palmitoylation of MPP1 (membrane-palmitoylated protein 1)/p55 is crucial for lateral membrane organization in erythroid cells. J Biol Chem. 2012;287:18974–18984. doi: 10.1074/jbc.M111.332981. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 145.Podkalicka J, Biernatowska A, Majkowski M, Grzybek M, Sikorski AF. MPP1 as a Factor Regulating Phase Separation in Giant Plasma Membrane-Derived Vesicles. Biophys J. 2015;108:2201–2211. doi: 10.1016/j.bpj.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Heberle FA, et al. Bilayer thickness mismatch controls domain size in model membranes. J Am Chem Soc. 2013;135:6853–6859. doi: 10.1021/ja3113615. [DOI] [PubMed] [Google Scholar]
- 147.Jensen MO, Mouritsen OG. Lipids do influence protein function – the hydrophobic matching hypothesis revisited. Biochimica Et Biophysica Acta-Biomembranes. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 148.Kaiser HJ, et al. Lateral sorting in model membranes by cholesterol-mediated hydrophobic matching. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16628–16633. doi: 10.1073/pnas.1103742108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Koster DV, Mayor S. Cortical actin and the plasma membrane: inextricably intertwined. Curr Opin Cell Biol. 2016;38:81–89. doi: 10.1016/j.ceb.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 150.Fritzsche M, Erlenkamper C, Moeendarbary E, Charras G, Kruse K. Actin kinetics shapes cortical network structure and mechanics. Science advances. 2016;2:e1501337. doi: 10.1126/sciadv.1501337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fujiwara TK, et al. Confined diffusion of transmembrane proteins and lipids induced by the same actin meshwork lining the plasma membrane. Mol Biol Cell. 2016;27:1101–1119. doi: 10.1091/mbc.E15-04-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Saha S, et al. Diffusion of GPI-anchored proteins is influenced by the activity of dynamic cortical actin. Mol Biol Cell. 2015;26:4033–4045. doi: 10.1091/mbc.E15-06-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Honigmann A, et al. A lipid bound actin meshwork organizes liquid phase separation in model membranes. eLife. 2014;3:e01671. doi: 10.7554/eLife.01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mueller V, et al. STED nanoscopy reveals molecular details of cholesterol- and cytoskeleton-modulated lipid interactions in living cells. Biophysical Journal. 2011;101:1651–1660. doi: 10.1016/j.bpj.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ehrig J, Petrov EP, Schwille P. Near-critical fluctuations and cytoskeleton-assisted phase separation lead to subdiffusion in cell membranes. Biophys J. 2011;100:80–89. doi: 10.1016/j.bpj.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Machta BB, Papanikolaou S, Sethna JP, Veatch SL. Minimal model of plasma membrane heterogeneity requires coupling cortical actin to criticality. Biophys J. 2011;100:1668–1677. doi: 10.1016/j.bpj.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu AP, Fletcher DA. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys J. 2006;91:4064–4070. doi: 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gowrishankar K, et al. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell. 2012;149:1353–1367. doi: 10.1016/j.cell.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 159.Koster DV, et al. Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer. Proc Natl Acad Sci U S A. 2016;113:E1645–1654. doi: 10.1073/pnas.1514030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fritzsche M, et al. Self-organizing actin patterns shape membrane architecture but not cell mechanics. Nature Communications. 2017 doi: 10.1038/ncomms14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rao M, Mayor S. Active organization of membrane constituents in living cells. Curr Opin Cell Biol. 2014;29:126–132. doi: 10.1016/j.ceb.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 162.Lingwood D, et al. Cholesterol modulates glycolipid conformation and receptor activity. Nature Chemical Biology. 2011;7:260–262. doi: 10.1038/nchembio.551. [DOI] [PubMed] [Google Scholar]
- 163.Filipp D, Leung BL, Zhang J, Veillette A, Julius M. Enrichment of lck in lipid rafts regulates colocalized fyn activation and the initiation of proximal signals through TCR alpha beta. Journal of immunology (Baltimore, Md: 1950) 2004;172:4266–4274. doi: 10.4049/jimmunol.172.7.4266. [DOI] [PubMed] [Google Scholar]
- 164.Zhang M, et al. CD45 signals outside of lipid rafts to promote ERK activation, synaptic raft clustering, and IL-2 production. Journal of immunology (Baltimore, Md: 1950) 2005;174:1479–1490. doi: 10.4049/jimmunol.174.3.1479. [DOI] [PubMed] [Google Scholar]
- 165.Field KA, Holowka D, Baird B. Fc epsilon RI-mediated recruitment of p53/56lyn to detergent-resistant membrane domains accompanies cellular signaling. Proc Natl Acad Sci U S A. 1995;92:9201–9205. doi: 10.1073/pnas.92.20.9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Varshney P, Yadav V, Saini N. Lipid rafts in immune signaling: Current progress and future perspective. Immunology. 2016 doi: 10.1111/imm.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dinic J, Riehl A, Adler J, Parmryd I. The T cell receptor resides in ordered plasma membrane nanodomains that aggregate upon patching of the receptor. Scientific reports. 2015;5:10082. doi: 10.1038/srep10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Beck-Garcia K, et al. Nanoclusters of the resting T cell antigen receptor (TCR) localize to non-raft domains. Biochim Biophys Acta. 2015;1853:802–809. doi: 10.1016/j.bbamcr.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 169.Sproul TW, Malapati S, Kim J, Pierce SK. Cutting edge: B cell antigen receptor signaling occurs outside lipid rafts in immature B cells. Journal of immunology (Baltimore, Md: 1950) 2000;165:6020–6023. doi: 10.4049/jimmunol.165.11.6020. [DOI] [PubMed] [Google Scholar]
- 170.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 171.Beissert S, et al. Impaired cutaneous immune responses in Thy-1-deficient mice. Journal of immunology (Baltimore, Md: 1950) 1998;161:5296–5302. [PubMed] [Google Scholar]
- 172.Lorizate M, et al. Comparative lipidomics analysis of HIV-1 particles and their producer cell membrane in different cell lines. Cellular microbiology. 2013;15:292–304. doi: 10.1111/cmi.12101. [DOI] [PubMed] [Google Scholar]
- 173.Farnoud AM, Toledo AM, Konopka JB, Del Poeta M, London E. Raft-like membrane domains in pathogenic microorganisms. Current topics in membranes. 2015;75:233–268. doi: 10.1016/bs.ctm.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Iwabuchi K. Involvement of glycosphingolipid-enriched lipid rafts in inflammatory responses. Frontiers in bioscience (Landmark edition) 2015;20:325–334. doi: 10.2741/4312. [DOI] [PubMed] [Google Scholar]
- 175.Teissier E, Pecheur EI. Lipids as modulators of membrane fusion mediated by viral fusion proteins. European biophysics journal: EBJ. 2007;36:887–899. doi: 10.1007/s00249-007-0201-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dick RA, Goh SL, Feigenson GW, Vogt VM. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc Natl Acad Sci U S A. 2012;109:18761–18766. doi: 10.1073/pnas.1209408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Staubach S, Razawi H, Hanisch FG. Proteomics of MUC1-containing lipid rafts from plasma membranes and exosomes of human breast carcinoma cells MCF-7. Proteomics. 2009;9:2820–2835. doi: 10.1002/pmic.200800793. [DOI] [PubMed] [Google Scholar]
- 178.Raghu H, et al. Localization of uPAR and MMP-9 in lipid rafts is critical for migration, invasion and angiogenesis in human breast cancer cells. BMC cancer. 2010;10:647. doi: 10.1186/1471-2407-10-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Larsen JB, et al. Membrane curvature enables N-Ras lipid anchor sorting to liquid-ordered membrane phases. Nat Chem Biol. 2015 doi: 10.1038/nchembio.1733. [DOI] [PubMed] [Google Scholar]
- 180.Cuesta-Marban A, et al. Drug uptake, lipid rafts, and vesicle trafficking modulate resistance to an anticancer lysophosphatidylcholine analogue in yeast. J Biol Chem. 2013;288:8405–8418. doi: 10.1074/jbc.M112.425769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gajate C, Mollinedo F. Edelfosine and perifosine induce selective apoptosis in multiple myeloma by recruitment of death receptors and downstream signaling molecules into lipid rafts. Blood. 2007;109:711–719. doi: 10.1182/blood-2006-04-016824. [DOI] [PubMed] [Google Scholar]
- 182.Shashkin P, Dragulev B, Ley K. Macrophage differentiation to foam cells. Current pharmaceutical design. 2005;11:3061–3072. doi: 10.2174/1381612054865064. [DOI] [PubMed] [Google Scholar]
- 183.Rios FJ, et al. Uptake of oxLDL and IL-10 production by macrophages requires PAFR and CD36 recruitment into the same lipid rafts. PLoS One. 2013;8:e76893. doi: 10.1371/journal.pone.0076893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Maguy A, Hebert TE, Nattel S. Involvement of lipid rafts and caveolae in cardiac ion channel function. Cardiovascular research. 2006;69:798–807. doi: 10.1016/j.cardiores.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 185.van Zanten TS, Mayor S. Current approaches to studying membrane organization. F1000Research. 2015;4 doi: 10.12688/f1000research.6868.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Toulmay A, Prinz WA. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. Journal of Cell Biology. 2013;202:35–44. doi: 10.1083/jcb.201301039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kahya N, Scherfeld D, Bacia K, Schwille P. Lipid domain formation and dynamics in giant unilamellar vesicles explored by fluorescence correlation spectroscopy. Journal of Structural Biology. 2004;147:77–89. doi: 10.1016/j.jsb.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 188.Kaiser HJ, et al. Order of lipid phases in model and plasma membranes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shogomori H, et al. Palmitoylation and intracellular domain interactions both contribute to raft targeting of linker for activation of T cells. J Biol Chem. 2005;280:18931–18942. doi: 10.1074/jbc.M500247200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


