Figure 2. Editing of wild-type human CD34+ HSPCs by the Cas9 RNP.
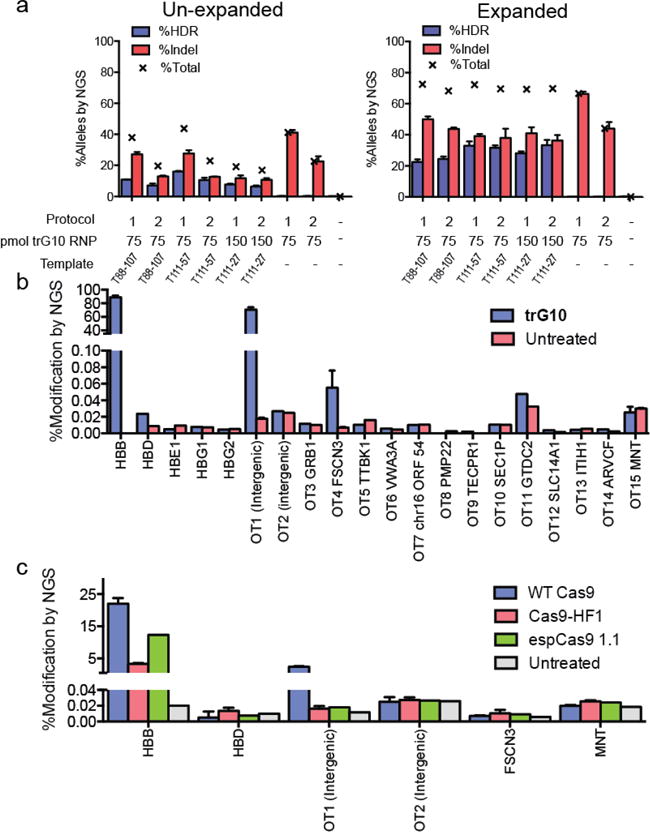
A) Analysis of editing in un-expanded HSPCs (left) and erythroid-expanded HSPCs (right), using trG10 RNP and conditions as indicated. Templates, which are asymmetric about the G10 cut site, were designed as described in the text. All samples are 3 biological replicates, error bars are ± standard deviation. B) Modification (HDR+Indel) at off-target sites in HSPCs edited with the trG10 RNP and template T88-107, compared to untreated cells. Samples for HBB, OT1, FSCN3, and MNT are from 3 biological replicates, with error bars ± standard deviation. Targets were selected using the online CRISPR-design tool (reference). C) Indel formation at on-and off-target sites by Cas9 mutants with increased specificity (HF1 and espCas9 1.1), compared to WT Cas9, all complexed to the G10 sgRNA and no ssODN. All samples are n=3 biological replicates, with error bars ± standard deviation.
