Abstract
To investigate the genetic basis of hereditary lens opacities we analyzed 31 cases of bilateral congenital cataract in Red Holstein Friesian cattle. A genome-wide association study revealed a significant association on bovine chromosome 7 at positions 6,166,179 and 12,429,691. Whole genome re-sequencing of one case and four relatives showed a nonsense mutation (g.5995966C>T) in the PZP-like, alpha-2-macroglobulin domain containing 8 (CPAMD8) gene leading to a premature stop codon (CPAMD8 p.Gln74*) associated with cataract development in cattle. With immunohistochemistry we confirmed a physiological expression of CPAMD8 in the ciliary body epithelium of the eye in unaffected cattle, while the protein was not detectable in the ciliary body of cattle with cataracts. RNA expression of CPAMD8 was detected in healthy adult, fetal and cataractous lenses.
Introduction
Cataracts are opacities of the lens present from birth (congenital) or acquired during life and are the main cause of blindness in humans worldwide [1]. The genetic background of the disease is well studied in humans. Until today more than 290 genes and 19 non-gene loci have been associated with cataract development in humans and mice [2]. Besides hereditary cataracts, opacities also occur as part of multisystemic disorders or due to the impact of environmental factors [3]. In cattle knowledge about the etiology of cataract development (due to genetic or environmental factors) is still relatively scarce, even though an incidence of 26% was reported in some herds [4]. Cataracts have already been observed in several cattle breeds, such as Holstein Friesian [5–8], Jersey [9, 10], Hereford [11], Aberdeen Angus [11], Shorthorn [11] and Ayrshire [4]. So far, only one mutation with recessive inheritance has been identified leading to juvenile-onset bilateral incomplete immature nuclear cataract in Romagnola cattle [12].
Many of the identified mutations leading to cataract development in humans and mice affect genes encoding lens crystallins and have been discussed in detail elsewhere [13]. Mutations in other proteins like membrane, cytoskeleton and gap junction proteins, beaded filaments, growth and transcriptional factors are also known to result in cataract development [3, 14]. Recently, C3 and PZP-like, alpha-2-macroglobulin domain-containing 8 gene (CPAMD8), a so-far unknown candidate gene for cataract development, was associated with anterior segment dysgenesis (ASD). ASD manifests itself in a spectrum of developmental abnormalities affecting the anterior segment of the eye, including cataract [15]. CPAMD8 belongs to the complement component 3 (C3)/alpha2-macroglobulin (A2M) family [16], whose members are involved in the innate immune system [17, 18]. Except for the association with ASD, knowledge about the biological function of CPAMD8 is comparatively scanty. The gene was discussed as being associated with Ashkenazi Jewish Crohn’s disease [19] and multiple sclerosis [20] in humans. Regarding eye development, CPAMD8 expression was observed to be upregulated in the regeneration-incompetent irides of axolotls [21].
Since 2009, congenital cataracts have been increasingly reported in German Red Holstein Friesian (HF) cattle. A total of 31 cases were observed, and they have provided the opportunity to elucidate the molecular cause of the disorder. As a result we have detected a nonsense mutation in CPAMD8 (g.5995966C>T) leading to a premature stop codon in exon 1 of the gene associated with congenital cataract development in HF cattle.
Results
Clinical findings
All four examined cases showed bilateral complete mature cataracts at time of birth. No other obvious ophthalmological anomalies were observed at this timepoint.
Eyes of the affected female cattle were examined in detail with the appropriate ophthalmologic equipment at approximately either 13 or 30 months of age (S1 Table). All cases showed a mature to hypermature cataract at time of examination (Fig 1A–1E). Besides the opacities, mydriasis and microphakia (mostly accompanied by an irregular shape of the lens), multiple eye defects like posterior synechia, glaucoma, exophthalmos, uveitis and retinitis (in one case) were additionally observed in some eyes (for details see S1 Table). These anomalies developed during life of cataract cases and were not congenital. The ongoing dissolution of the lens and subsequent inflammatory processes affecting the eye are suspected to be the reason for these clinical findings. In all but one eye the ocular fundus appeared to be free of pathological findings. Anomalies as iris hypoplasia, corectopia and ectropion uveae as observed by Cheong et al. (2016) were not detected. Neurological examination did not show any deficits other than the ones related to the bilateral eye problem.
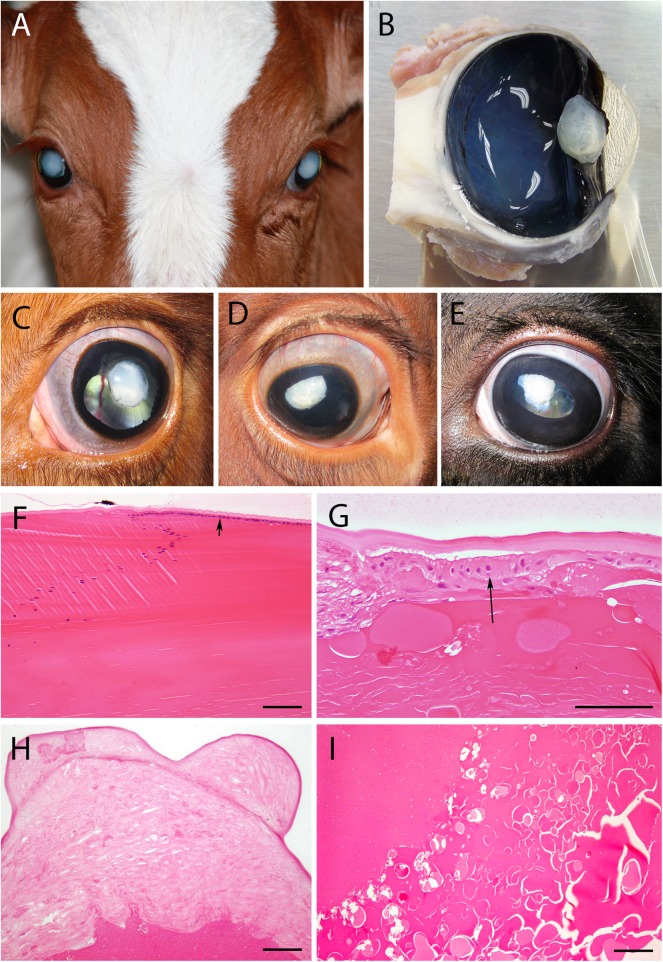
Fig 1. Congenital cataract in Red Holstein Friesian cattle.
(A) Bilateral mature cataract formation, (B) sagittal cut through the eyeball (formalin fixed) with cataractous lens (animal #224) clearly showing the irregular lens surface. (C) to (E) are pictures of different stages of the degenerative lenses in situ taken during the clinical examinations: (C) left eye of animal #227 at 13 months, mature cataract, (D) right eye of animal #489 at 31.5 months, mature to hypermature cataract, (E) left eye of animal #908 at 30 months, mature to hypermature cataract. Iris abnormalities as observed by Cheong et al. (2016) in human ASD patients showing cataracts were not detected in bovine cataract cases. (F) to (I) show the histopathological changes in hematoxylin/eosin stained sections (bars = 100μm): (F) lens equatorial line in a bovine healthy adult lens where the anterior epithelium (arrow) under the capsule is clearly visible (control animal). During cataract formation the capsule thickens, the cuboidal epithelium (G, arrow, animal #224, left eye) disintegrates and over time vanishes completely (H, animal #908, left eye). The loss of lens fibrils and presence of Morgagnian globules indicate the presence of a hypermature cataract (G, I, animal #224). Fig 1A was digitally improved and cropped using GIMP 2.8.18.
Pathology and histopathology
Since the complete eyeballs were fixed in formalin before they were cut for histology, the visual inspection of the inner eye was performed on fixed tissue. Lenses were usually of irregular shape, about two-thirds to half the size of an unaffected lens, and opaque white (Fig 1A–1E). Microphakia was more pronounced in two of the older animals (#908 (Fig 1E) and #489 (Fig 1D), 30 and 31.5 months of age). Histologically, the regular lens would consist of a cuboidal epithelium in its anterior parts (Fig 1F arrow) that turns at the equatorial line, loses its nuclei and cytoplasmatic organelles and degrades to lens fibers, building the bulk of the lens. The lens epithelium usually has a thick basement membrane, forming a capsule (Fig 1F). The cataractous lenses showed a loss of lens epithelium and thickening of the capsule (Fig 1G and 1H), but no clear distinction between basement membrane and connective tissue. The bulk of the lens showed Morgagnian globules (Fig 1G and 1I), liquefactions and mineralizations (not shown) confirming the presence of a hypermature cataract. No fibrillary structures could be observed.
Autosomal recessive inheritance of congenital cataract formation
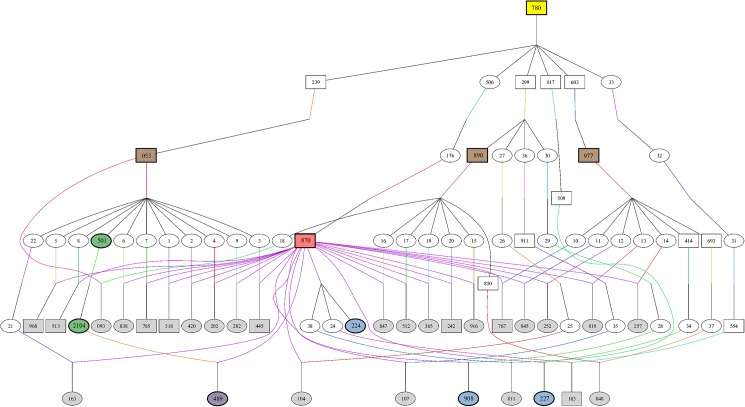
Analysis of the pedigree data revealed that 26 of 31 cases were paternal half-siblings of sire #870 (Fig 2). In 20 of 31 cases, the dams’ fathers were either sires #053, #890 or #977. The four sires #870, #053, #890 and #977 shared one common ancestor (#780) two to three generations before. Sire #780, born in 1982, appeared in 28 of 31 cases in the dam and sire line of the pedigrees. Pedigrees of the remaining three animals were not clearly documented. Parental testing was not performed. However, an autosomal recessive inheritance of the cataract phenotype can be inferred from pedigree analyses, although a relation to the predicted founder (#780) on the dam´s side could not be verified in three cases. Sire #780 was genotyped as a carrier and is most likely the founder of the defect.
Fig 2. Origin and transmission of cataract in Red Holstein Friesian cattle.
The pedigree depicts the ancestry of the affected individuals (grey, purple and blue symbols). Two cases are not displayed in the family tree. Neither maternal nor paternal lines were clearly documented. Animal #183 was also genotyped T/T for the variant g.5995966C>T in CPAMD8. The origin of the cataract-causing mutation was traced back to founder sire #780 (yellow symbol). The dam´s sires in 20 of 31 cases were sires #053, #890, #977, marked with brown symbols. The red symbol shows sire #870, father of 26 of 31 cases. This animal (#870) was also used for whole genome re-sequencing, as the green (mother and maternal grandmother of one case) and purple (cattle with congenital cataract) marked cattle. The blue and purple symbols mark the four dissected cases. Family tree was created using Pedigraph [22].
Association and region of extended homozygosity on bovine chromosome 7 (BTA7)
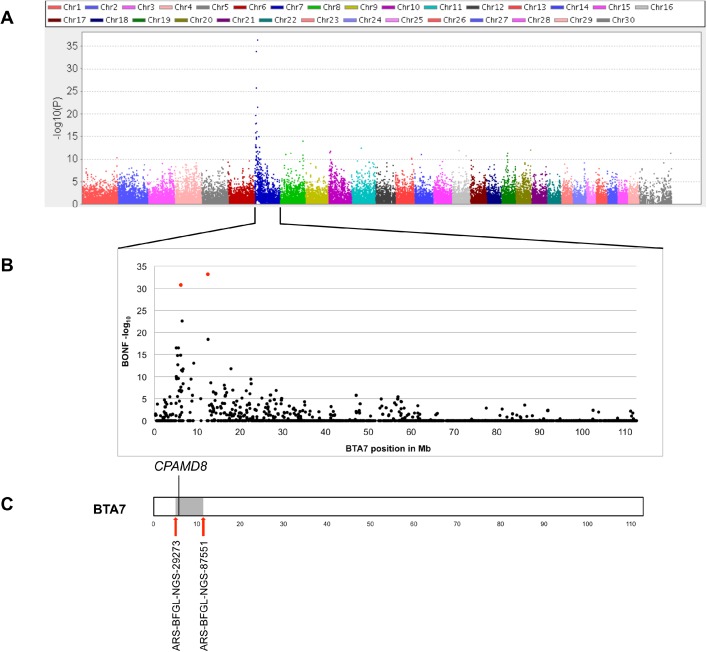
Case-control analysis of 26 cases and 88 controls revealed an association with the cataract phenotype on bovine chromosome 7 (BTA7) (p values: 4.41x10-37 (Bonferroni–adjusted p value: 1.85x10-32) at position 12,429,691 and 1.30x10-34 (Bonferroni–adjusted p value: 5.44x10-30) at position 6,166,179) (all positions refer to assembly UMD_3.1) (Fig 3). Regarding the proposed autosomal recessive inheritance, we searched for regions of extended homozygosity in cases of congenital cataracts. A 4.7 Mb interval from 5,639,104 to 10,406,009 on BTA7 (UMD_3.1) was detected harboring 76 SNPs. Due to the positional overlap of the association on BTA7 and the region of homozygosity on BTA7, we searched for positional and functional candidate genes within this region. However, none of the 166 annotated genes located in the region of extended homozygosity had so far been associated with congenital cataract.
Fig 3. Results of genome-wide association study (GWAS) using SNP data of 26 cases and 88 Holstein control animals.
(A) Manhattan plot (generated using Haploview [23]) and calculated -log10 p values showing an association with the cataract phenotype on BTA7, (B) Detailed view of association on BTA7 with -log10 Bonferroni-adjusted p values. Red symbols showing associations of 5.44x10-30 at position 6,166,179 and 1.85x10-32 at position 12,429,691, (C) Region of extended homozygosity on BTA7 in cataract cases (grey symbol). CPAMD8 (5,995,747 to 6,095,877) is located at the proximal end of the 4.7 Mb region of extended homozygosity from 5,639,104 to 10,406,009.
Whole genome re-sequencing and identification of a variant in the CPAMD8 gene associated with congenital cataract formation in cattle
Due to the lack of functional candidate genes in the associated region on BTA7, a whole genome re-sequencing was performed. Based on the pedigree analysis two trios were sequenced. The first trio consisted of one case (#489, Fig 1D) and its parents (#870 and #2104). The second trio included the grandmother (#501) and a cross between the grandmother #501 and the father #870 of this case #489. 2019 variants in the homozygous state were called for the affected animal in the associated interval from 4 to 13 Mb on BTA7. The detected variants were filtered for their predicted effect on the amino acid sequence. Seven variants with non-synonymous coding and one variant leading to a premature stop codon remained after filtering. Six of seven variants with non-synonymous coding affected olfactory receptors (olfactory receptor 10H1-like, 10H1 and 10H4) and were excluded from further analysis due to their biological function. Remaining variants in GTPBP3 (GTP binding protein 3, g.5711730T>C) and CPAMD8 (C3 and PZP like, alpha-2-macroglobulin domain containing 8, g.5995966C>T; positions refer to accession number AC_000164.1, UMD_3.1) were validated by PCR and subsequent melting curve analysis using FRET technology. The only variant that remained perfectly associated with the cataract phenotype was the chain termination mutation detected in CPAMD8 (g.5995966C>T). CPAMD8 is located from 5,995,747 to 6,095,877 on BTA7 (UMD_3.1, AC_000164.1) and contains 42 exons. The gene is located at the proximal end of the detected region of extended homozygosity on BTA7 and at a distance of around 70 kb from a SNP at position 6,166,179 (Bonferroni-adjusted p value of 5.44x10-30, UMD_3.1, AC_000164.1), which was identified previously by GWAS. The identified variant g.5995966C>T was located within the first exon of the CPAMD8 gene at position c.220 (UMD_3.1) and has not yet been described. The nonsense mutation p.Gln74* results in a null allele. Homozygous mutant cases therefore lack CPAMD8 protein.
Genotyping of variant g.5995966C>T
Genotyping of 1,248 animals was performed to verify the variant g.5995966C>T in CPAMD8 (Table 1). All animals with cataract phenotype (n = 31) were tested T/T. In total 161 cattle were genotyped C/T, including the parents of affected cattle. 1,055 randomly tested control animals were tested homozygous C/C. All pedigrees of heterozygous animals showed an inheritance with the predicted founder, sire #780.
Table 1. Detection of CPAMD8 g.5995966C>T in different cattle breeds.
a) The 32 T/T individuals include 31 cases and one HF animal with unknown phenotype (#183). Both parents of #183 were tested heterozygous C/T and were related to the predicted founder sire #780.
b) All heterozygous tested HF cattle were related to sire #780.
c) The 1,055 C/C individuals include 1,046 HF and nine animals of other breeds, i.e. Angus, Charolais, Jersey, Limousin, White Galloway.
Besides the 31 tested cataract cases, there was an additional animal (#183) genotyped T/T. Unfortunately, the phenotype of the animal was unknown, since it had already been culled by the time of genotyping. However, the father (#554) and the maternal grandfather (#870) of #183 were tested heterozygous C/T and were related to sire #780.
RNA expression of bovine CPAMD8
RNA was extracted from the lenses of three animals, one with cataract (mature to hypermature cataract, #489), one adult control and one fetal control animal. Furthermore, lung and thyroid gland tissues from two animals, one with cataract (#489) and one adult control animal, were used as control samples (Table 2). Other ocular tissue samples were not considered. Except for cataracts, observed ocular anomalies were not congenital. Anomalies as listed in S1 Table, developed due to ongoing lens dissolution and subsequent inflammatory processes affecting the eyes of cataract cases. Due to the physiological condition of the other ocular structures, these tissues were not considered for RNA analyses. Although RNA from cataractous and adult lens extracts showed low RNA quality (RIN factors <6), CPAMD8 RNA was detected in all tested tissue samples. However, an exact quantification relative to the reference gene was not possible. Despite the low quality, CPAMD8 RNA was detectable in lens materials of cataractous, fetal and healthy adult lenses.
Table 2. Tissue samples for RNA analyses of CPAMD8, including ddPCR results and RNA integrity number (RIN) from RNA quality measurement using RNA 6000 Pico Kit (Agilent Technologies, Waldbronn, Germany).
| Tissue samples | RIN | Mean copy numbers CPAMD8 (CV1) | Mean copy numbers GAPDH (CV1) |
|---|---|---|---|
| Lung-healthy control | 7.4 | 23.3 (0.107) | 99 (0.0808) |
| Thyroid gland-healthy control | 6.9 | 11.5 (0.260) | 1442.5 (0.0947) |
| Lung case | 8.3 | 59.5 (0.032) | 127 (0.0394) |
| Thyroid gland case | 7.4 | 8.2 (0.012) | 228 (0.0263) |
| Adult healthy lens | 2.4 | 0.125 (0.360) | 120.5 (0.0622) |
| Fetal lens | 7.1 | 0.665 (0.0226) | 597.5 (0.0410) |
| Cataractous lens | 1 | 0.023 (0.231) | 0.32 (0.0625) |
1) CV = Coefficient of variation
Annotation of CPAMD8 is based on assembly UMD_3.1, AC_000164.1. Expression of corresponding isoforms was tested by PCR for lens tissue. There was no indication of expression of alternative isoforms as e.g. ENSBTAP00000036263, annotated in Ensembl database.
Immunohistochemical detection of CPAMD8
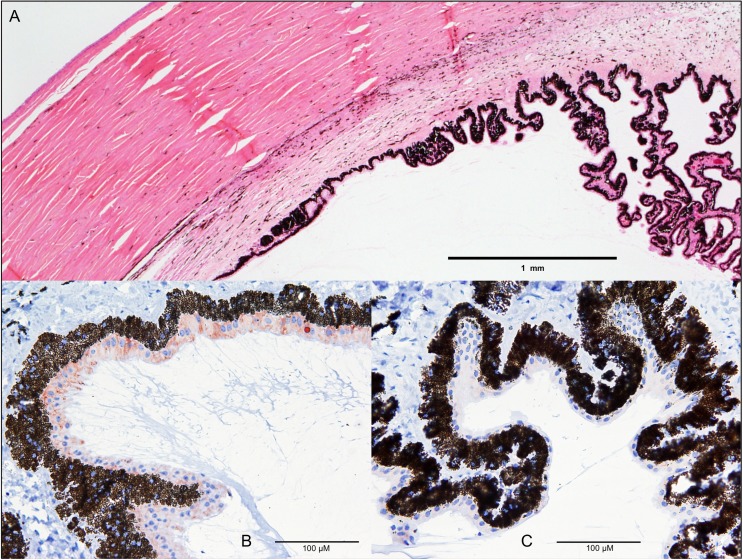
In healthy fetal and adult control cattle, CPAMD8 was detectable by immunohistochemistry in the epithelium of the ciliary body, where the staining was related to intracytoplasmic organelles (Fig 4B). In contrast, in the ciliary body epithelium of cataract cases, no CPAMD8 immunoreaction was detectable (Fig 4C). In cataract and control cases alike the retina, the epithelium of the lens and the lentil fibers showed no antibody reaction for CPAMD8 in the immunohistochemical staining. However, using Western blotting a faint band at the expected size of CPAMD8 was detectable in normal fetal and normal adult but not adult cataractous lenses (S1 Fig).
Fig 4. Loss of CPAMD8 expression in cataract cases.
Overview of cornea and ciliary body (A, H&E staining). Physiologically, CPAMD8 is detectable by immunohistochemical staining in the epithelium of the ciliary body as cytoplasmic and vesicular staining in light brown (adult control sample) (B). In the cataract ciliary body epithelium, no CPAMD8 immunoreaction is detectable (C); CPAMD8 antibody; visualization by AEC.
Discussion
In the present study we identified a chain termination mutation (g.5995966C>T) in the first exon of CPAMD8 associated with congenital cataract formation in Red Holstein Friesian cattle. Information about the potential biological function of CPAMD8 is relatively scarce. Recently, Cheong et al. (2016) identified several mutations (missense, splice-site and frameshift variants) in CPAMD8 associated with ASD in humans, a disorder causing malformations of the anterior segment of the eye [15]. Affected patients showed several iridal malformations and cataracts, mainly of the posterior cortical type [15]. Observed bovine cataract cases with congenital mature cataracts did not show any other congenital anomalies of the iris or other ocular tissues. Likewise identified mutations in ASD patients and cataract cattle observed here differed regarding location and effects on the protein. Cheong et al. (2016) identified a homozygous missense mutation (c.4351T>C, p.S1451P) in exon 32 in one family affecting the conserved A2M domain of the CPAMD8 protein. In two other families two compound heterozygous mutations were identified respectively, causing aberrant pre-mRNA splicing resulting in an in frame deletion/insertion or smaller transcripts. The identified mutation c.2352_2353insC in family 2 is predicted to cause a premature termination in exon 18 of 42 resulting in a truncated protein [15]. Here a missense mutation (g.5995966C>T) in exon 1 of bovine CPAMD8 gene is reported leading to a premature termination that is predicted to cause a loss of protein in homozygous affected animals. In contrast, findings of Cheong et al. (2016) showed that a truncation of the protein by missense mutation, compound heterozygous frameshift mutations or aberrant pre-mRNA splicing are causing malformations of the anterior segment of the eye including cataracts.
Furthermore, we detected CPAMD8 in the ciliary body epithelium of healthy fetal and adult cattle, whereas no expression was detected in cataract cases. The non-pigmented part of the ciliary body secretes the aqueous humor by a complex process including steps of diffusion and ultrafiltration [24, 25]. Proteins in aqueous humor, although the concentration is rather low [25], are thought to derive from several sources like the plasma and the ciliary body itself [26, 27]. Goel et al. (2013) identified 2,815 proteins in the ciliary body, 896 of which are unique to the tissue and cannot be found in the plasma [27]. Due to our observation of CPAMD8 in the non-pigmented epithelium of the ciliary body of healthy cattle, we assume that the CPAMD8 protein is usually secreted into the aqueous humor in the physiological state. This is supported by previous reports of CPAMD8 protein in human ciliary body [27] and aqueous humor [28–30]. To the best of our knowledge there have been no reports about defects of any other ciliary body proteins causing cataracts so far. CPAMD8 was not detectable by immunohistochemistry in lens tissues of fetal (approximately 11 weeks of age) and adult controls as well as cataract animals. However, the presumed absence of CPAMD8 in fetal and normal adult lenses was most likely only due to the reduced intensity of the immunohistochemical staining. Because, when using Western blotting as a more robust and quantitative method [31], a faint band at the expected size of CPAMD8 was also detectable in normal fetal and adult but not cataractous lenses (S1 Fig). So far there have been no reports of CPAMD8 in mammalian lens tissues, although it was detected in other ocular tissues and fluids e.g. aqueous humor [28–30], ciliary body [27], zonular fibres [32], vitreous humor [33], iris [34, 35], and choroid-retinal pigment epithelial complex [36].
We investigated CPAMD8 RNA expression in several bovine tissues including adult, fetal and cataractous lenses. Likewise, Cheong et al. (2016) reported an expression of CPAMD8 in human fetal lens in increasing levels from week 9 to 22 of gestation [15], at a time when the first lens fiber cells are already being formed (by the seventh week of gestation) [37]. Due to the similar pregnancy periods of humans and cattle, this clearly corresponds with our findings of CPAMD8 expression in bovine fetal lens tissue derived from a fetus of approximately 10 weeks of age according to the measured crown-rump length. Furthermore, CPAMD8 expression was detected in adult and cataractous lens material, indicating a postpartal expression of CPAMD8. Due to degradation in cataractous lenses RNA quantification was not possible.
Nevertheless, our findings of CPAMD8 RNA expression in lens tissues and CPAMD8 protein in ciliary body and lenses of healthy cattle are in accordance with previous reports [15, 27]. In which way lens development might be influenced by the loss of CPAMD8 protein is a question that requires further investigation.
Concerning the structure of CPAMD8, the protein belongs to the C3/Α2M family, which has a further six members, including A2M, PZP, CD109 and the complement components C3, C4, C5 [16]. The CPAMD8 gene has some structural similarities in common with other family members like, for example, A2M-associated domains and a Kazal-type serine protease inhibitor/follistatin-like domain, which is highly conserved across vertebrate species [16].
Functionally, members of the C3/A2M family are part of the innate immune system [17, 18]. Complement components recognize and eliminate pathogens, i.e. elimination is achieved by opsonization, lysis of pathogens and inflammatory response [18]. Alpha-2-macroglobulin (Α2M) proteins are able to capture proteases of endogenous and exogenous origin and are evolutionarily highly conserved elements [17]. Ji et al. (2016) identified proteomic changes of the complement components C3 and C5 in aqueous humor of cataract patients and controls, being associated with cataract development.
Seeking a potential role for CPAMD8 during cataract development, structurally similar Α2M was identified acting as a carrier of specific growth factors, like as e.g. TGFβ [38]. TGFβ family members are present in ocular surroundings, i.e. lens [39, 40], aqueous and vitreous humor [30, 33, 41]. Changes in TGFβ levels are known to result in cataract development [42], indicating a strong regulation of the expression and bioavailability of TGFβ. Also, TGFβ signaling is thought to have a key role during terminal fiber cell differentiation [43]. It has already been shown that Α2M in vitro, when present in ocular media, blocks cataractous changes induced by TGFß [44], suggesting that suitable levels of molecules like A2M in the ocular media have a protective function for lens cells against the damaging effects of TGFß. Here, reduced levels of A2M or similar molecules may predispose the lens to cataractogenesis. However, cataractous changes induced by TGFß are usually similar to those found in anterior subcapsular cataracts [45]. ASD patients with mutations in CPAMD8 mainly suffer from cortical [15] and affected cattle from complete mature cataracts. These differences in cataract manifestation may indicate that CPAMD8 and Α2M, in terms of cataract prevention, act by different pathophysiological mechanisms, although they have structural similarities and have expression in aqueous humor and ciliary body in common.
Clinical examination of affected cattle in this study revealed a mature cataract at time of birth. With age, the cataract manifestation changed from mature to hypermature. Histological results, such as Morgagnian globules, liquefaction, the absence of lens epithelium and reduction of lens size, are typical for hypermature cataract progression of cortical cataracts [46]. Alterations leading to congenital, mature cataracts most likely affect a stage in embryonic lens development when lens fiber cells are already formed. Affected cattle seemed to have normal-sized lenses at the time of birth, which supports this hypothesis.
Besides the inherited form of cataractogenesis, intrauterine infections play a crucial role in cataract development. Bovine Viral Diarrhea-Mucosal Disease (BVD-MD) is known to be responsible for the development of cataracts in cattle [47]. Twenty-two of the 31 sampled cataract cases were tested for BVD-MD by antigen-ELISA test with negative results. Therefore, an intrauterine infection as the cause of the cataracts could be excluded.
Our findings are not only interesting from a scientific point of view, they also have substantial relevance for practical breeding. Genotyping of male offspring of identified carriers revealed further heterozygous sires. Two of these tested heterozygous males were ranked top ten in the active, daughter-proven, artificially inseminated German Red Holstein population (ranked by relative breeding value) in August 2015 [48].
In summary, we have discovered a nonsense mutation (g.5995966C>T, p.Gln74*) in the bovine CPAMD8 gene which coincides perfectly with cataract in the tested animals. We demonstrated that CPAMD8 is expressed in the ciliary body epithelium and lens of the unaffected eye and assume that a secretion of CPAMD8 from the epithelium into aqueous humor takes place under normal circumstances. The question of how in detail the loss of CPAMD8 in these cases results in cataract development needs to be examined in further studies. Nevertheless, our data provide convincing evidence that the absence of CPAMD8 leads to congenital cataract formation during embryonic development in Red Holstein Friesian cattle and are in accordance with previous findings that CPAMD8 is involved in eye development in humans [15].
Materials and methods
Animal ethics/ethics statement
The current study was performed in accordance with the ethical guidelines of the University of Goettingen and the German Animal Welfare act. Genotyping data and blood sampling had been done independent of the project in the frame of routine diagnostic parentage control and genomic selection programs of the German Holstein Friesian Association (DHV). The data sets were provided by DHV with written owner consent. Collection of blood samples within the breeding programs have been conducted exclusively by state-licensed veterinarians in accordance with the German Animal Welfare Act (§6 Abs. 1 Satz 2 TierSchG). Tissue samples and other native materials were obtained during or after regular slaughtering processes at an abattoir according to §4 of the German Animal Welfare Act. Therefore no formal ethical approval was required, since no other samples were collected for this study.
Animal material and sample collection
DNA samples from 31 affected (24 female, 7 male) Holstein Friesian cattle were available. Genomic DNA had been extracted from EDTA blood using a modified salting out procedure [49] or MagNa Pure LC DNA Isolation Kit I (Roche Diagnostics, Mannheim, Germany). DNA of related sires and other animals in the study was extracted from semen using a modified salting out method or obtained from the DNA depository of the Institute of Veterinary Medicine (Goettingen). Sample material for whole genome re-sequencing was extracted from EDTA blood as described above or semen using a phenol chloroform extraction method [50]. Lenses were obtained during regular slaughtering and stored at -80°C or fixed in 10% neutral buffered formalin for histological analysis. Fetal lenses were prepared from fetus (approximately 79 (immunohistochemistry) and 69 (RNA analysis) days of age according to measured crown-rump length) of dam during regular slaughtering and handled as described above.
Genome-wide association study (GWAS) and homozygosity mapping
Genotyping was performed using the BovineSNP50 BeadChip (Illumina, San Diego, USA) according to the manufacturer´s protocols. In total, 26 affected and 88 control HF cattle were genotyped. Final reports were generated using GenomeStudio V2011.1 (Illumina, San Diego, USA). Statistical analyses were performed using PLINK software [51]. Genotype data were filtered for quality control criteria as SNP call rate >95% and minor allele frequency ≥ 5%, reducing the final data set to 46,075 SNPs.
Firstly, a case-control association was calculated. P values were Bonferroni-adjusted to correct for multiple testing. Regions of extended homozygosity with shared alleles in cases were analyzed in a second step. Regions larger than 1 Mb were included in the analysis. Genomic positions refer to assembly UMD_3.1.
Genome re-sequencing and variant calling
Whole genome shotgun sequencing was conducted on five cattle: one affected animal (#489) and its unaffected parents (#870 and #2104), one unaffected maternal grandmother (#501) and an unaffected cross between the maternal grandmother (#501) and the father of the affected cattle (#870). Sequences were deposited with the European Nucleotide Archive (ENA) under accession number PRJEB20549 (http://www.ebi.uk/ena/data/view/PRJEB20549).
Sequencing libraries were prepared from 100ng DNA using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, USA) according to the manufacturer’s instructions. Paired-end sequencing was conducted on a NextSeq500 (Illumina, San Diego, USA).
An average of 85M (SD: 7.8M) reads were generated per animal, of which 83% (SD: 1.3%) were mapped to the bovine reference genome (UMD_3.1) resulting in a mean coverage of 7.8-fold (SD: 0.78).
Sequences were mapped using a BWA software package [52]. Variants were named using GATK [53]. Effects on the amino acid sequence were predicted by the SNPeff tool [54].
Genotyping by PCR using FRET technology
SNP genotyping was done by fluorescence resonance energy transfer-PCR [55]. Primers were designed using NCBI primer-BLAST [56] with the following sequences: CPAMD8_FRET_93bp_F: GTTCTTGCTGCTGCTGCTG and CPAMD8_FRET_93bp_R: CAGCCAGCCTTCTCTCGC. Probes, bCPAMD8_probe: Cy5-GCGCTGAGCAGCCCCA-PHO and bCPAMD8_anchor: CGCCGCGGGACGGCG-Flc, were designed using the MeltCalc software [57, 58]. Primers and probes were synthesized by Sigma-Aldrich (Taufkirchen, Germany).
PCR was performed on a LightCycler480 or LightCycler480 II (Roche Diagnostics, Mannheim, Germany) in a total volume of 25μl using FastStart Taq DNA Polymerase, dNTPack (Roche Diagnostics, Mannheim, Germany). One reaction mix included 1.5U Faststart Taq DNA Polymerase, 200μM dNTP, 10μM of each primer and probe, 1x GC-RICH solution, 4.5mM MgCl2, 1x PCR reaction buffer (including 20mM MgCl2), and 60-150ng of DNA. Cycling conditions were 95°C for 10 min, followed by 35 to 40 cycles of 95°C for 10 sec, 59°C for 20 sec and 72°C for 10 sec. Final elongation step was 72°C for 5 min. Melting was done using an appropriate filter set and the following program: 95°C for 30 sec, 45°C for 30 sec, 75°C continuous acquisition mode (2 to 3/°C), ramp rate 0,29°C/sec, followed by 45°C for 30 sec.
RNA extraction and cDNA product amplification
RNA was extracted from 50 to 100 mg tissue material homogenized in 900 μl TRIzol® (Thermo Fisher Scientific) using Magna Lyzer Green Beads (Roche Diagnostics, Mannheim, Germany). After homogenization, RNeasy Plus Universal Kit (Qiagen, Hilden, Germany) was used following the manufacturer´s protocol. RNA quality control was tested using Agilent RNA 6000 Pico Kit (Agilent Technologies, Waldbronn, Germany). cDNA synthesis was performed using Maxima H Minus First Strand cDNA Synthesis Kit, with dsDNase (Life Technologies). Oligo(dt) and Random Hexamer primers were used for reverse transcription.
Digital droplet PCR assay for RNA expression analysis
CPAMD8 RNA expression analyses were performed on a QX200Droplet Digital PCR system (Bio-Rad Laboratories, Munich, Germany) using RNA samples of one cataract case, one healthy adult and one fetal control. One technical replication was performed for every sample. Primers were designed using NCBI primer-BLAST [56]. Sequences of the primers were: CPAMD8_F: GATGGGAAGTCCGTCAGACC and CPAMD8_R: TGGACTCTCTCGTTGGGACA. As controls served ACTH and GAPDH with the following primer sequences: ACTB_cDNA_F: ACAGGATGCAGAAAGAGATCAATG, ACTB_cDNA_R: GTACTCCTGCTTGCTGATCCAC, GAPDH_cDNA_F: GTATGATTCCACCCACGGCA and GAPDH_cDNA_F: ACCACATACTCAGCACCAGC. All primers were synthesized by Sigma-Aldrich (Taufkirchen, Germany).
PCR mix at a total volume of 20μl included 10μl of QX200™ ddPCR™ EvaGreen Supermix (Bio-Rad Laboratories, Munich, Germany) and 2 μM of each primer. 1 μl to 2 μl of diluted samples (CPAMD8 1:2, ACTH and GAPDH 1:250) were used in PCR. cDNA of cataract case was used without dilution (2μl). Droplets were generated using QX200 Droplet Generator (Bio-Rad Laboratories, Munich, Germany). PCR was performed in a thermocycler (Biometra, Göttingen, Germany) and cycling conditions were as follows: 95°C for 10 min, followed by 45 cycles of 95°C for 30 sec and 60°C for 1 min. Final droplet stabilization was performed at 98°C for 10 min. Droplets were analyzed using the QX200 Droplet Reader (Bio-Rad Laboratories, Munich, Germany).
Histology
Tissues were fixed in 10% neutral buffered formalin for at least 48 h, embedded in paraffin, and 1–3μm tissue sections were cut with a microtome. Sections were mounted on glass slides, dried and stored for histological and immunohistochemical staining procedures. The morphological evaluation was performed on hematoxylin and eosin (H&E) stained sections.
Immunohistochemistry and Western blotting
For protein analyses four commercially available antibodies were used (S2 Table). Specificity and sensitivity of the antibodies were tested using different bovine tissues and an in vitro expressed recombinant bovine CPAMD8 epitope (S1 Appendix, S2 Fig). Antibody concentrations were titrated until an optimal signal to noise ratio was obtained. For immunohistochemistry antibodies HPA031327 and HPA031328 were initially tested and HPA031328 gave best signals. Therefore, this antibody was used for the further studies. For immunohistochemistry paraffin-embedded tissue sections were deparaffinized and rinsed with TBS. For antigen epitope retrival, slides were heated in a microwave at 200W for 5 x 3 min in TE-buffer (10mM Tris, 1mM ETDA, pH 9.0) and unspecific antibody binding was blocked with 3% hydrogenperoxide and 10% donkey serum in TBS-Triton (50mM Tris, 150mM NaCl, 0.1% Triton X-100).
Immunoreaction was performed by incubation with the affinity purified polyclonal rabbit anti-CPAMD8 antibody (HPA031328, Sigma-Aldrich) (1:50, according to manufacturer’s instructions) diluted in TBS-Triton followed by the secondary donkey-derived anti-rabbit antibody (RPN1004, GE Healthcare, Hamburg, Germany) (1:100) for 2 h and 1 h, respectively. After each step, slides were rinsed thoroughly with TBS-Triton. The immunoreaction was visualized using Avidin-conjugated peroxidase as enhancer and AEC (3-Amino-9-ethylcarbazole) as chromogen. Sections were lightly counterstained with hematoxylin.
For Western blotting proteins were lysed using lysis buffer containing 50mM Tris, 150mM NaCl, 1mM EDTA, 1mM EGTA, 5% glycerol, 1% Triton X-100, 0.1% DTT and EDTA free protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Homogenization of samples was done using DIAX900 Homogenizer (Heidolph, Schwabach, Germany) or MagNA Lyser Green Beads (Roche Diagnostics, Mannheim, Germany) followed by an incubation for 1 hour and centrifugation for 10 min at 500g and 4°C. Proteins were quantified by Bradford method.
60μg of proteins were analyzed using SDS-Page (4–12% Bis Tris- Plus gels or 8% Bis Tris Plus gels, Life Technologies). As positive controls human CPAMD8 over-expression Lysate (OriGene Technologies, Rockville, USA) and 5μl of in vitro produced bovine CPAMD8 were used. Proteins were transferred to nitrocellulose membrane using semi-dry blotter (Brenzel Bioanalytik, Lahntal, Germany). After transfer membranes were blocked with 5% non-fat dry milk in TBS-T for 2 h, followed by incubation with anti-CPAMD8 antibody (S2 Table, dilution 1:200) for 1 h at room temperature. Secondary goat anti-rabbit IgG (H + L)-HRP conjugate antibody (Bio-Rad Laboratories, Munich, Germany) was diluted 1:5000 and also incubated for 1 h at room temperature. Detection of chemiluminescent signals was performed using Amersham ECL detection reagent (GE Healthcare, Hamburg, Germany) and membranes were exposed to X-ray films (GE Healthcare, Hamburg, Germany).
Supporting information
Protein extracts of a normal adult lens (lane 2), normal fetal lens (lane 3) and adult cataractous lens (lane 4) were loaded on a 4–12% gradient SDS-PAGE. As positive control human CPAMD8 over-expression lysate was used (lane 1). Proteins were transferred to nitrocellulose membranes and filters were blocked with 5% non-fat dry milk in TBS-T followed by incubation with anti-CPAMD8 antibody HPA031328 (S2 Table, dilution 1:200). Detection of chemiluminescent signals was performed using Amersham ECL detection reagent and membranes were exposed to X-ray films. CPAMD8 bands in lane 1 (human control lysate), 2 (bovine normal adult lens) and 3 (bovine normal fetal lens) are indicated with an arrow. Protein sizes are indicated in kDa. Scans of X-ray films were cropped using GIMP 2.8.18 (GNU Image Manipulation Program).
(TIFF)
Five μl of in vitro produced bovine CPAMD8 epitopes were loaded on a 4–12% gradient SDS-PAGE (lane 1, lane 2). Proteins were transferred to nitrocellulose membranes and filters were blocked with 5% non-fat dry milk in TBS-T followed by incubation with anti-CPAMD8 antibody HPA031328 (S2 Table, dilution 1:200). Detection of chemiluminescent signals was performed using Amersham ECL detection reagent and membranes were exposed to X-ray films. The band in lane 1 shows the expected size of the epitope (approx. 33 kDa) and is indicated with an arrow. In lane 2 another in vitro expressed epitope lysate was loaded showing no reactivity. Lane 3 shows a negative control lysate. Protein sizes are indicated in kDa. Scans of X-ray films were cropped using GIMP 2.8.18 (GNU Image Manipulation Program).
(TIFF)
Consciousness, posture, gait, swallowing and tongue tone were normal in all cases. Behavior differed from calm alertness to anxiety. If not stated otherwise the ocular fundus was without pathological findings.
(DOCX)
1) Sigma-Aldrich Chemie (Taufkirchen, Germany); 2) OriGene Technologies (Rockville, USA).
(DOCX)
(DOCX)
Acknowledgments
The authors thank S. Pach, R. Andag, M. Scharfenstein, S. Loos, L. Jüttner, S. Bierau, S. Balzer and K. Brekerbohm for their excellent technical assistance. We are grateful to T. Leeb who kindly validated our initial PLINK-data. J. Hartmann (Rinder-Union West eG) is thanked for providing samples and data. Thanks to H. Melbaum for providing semen samples. F. Reinhardt (VIT) and D. Segelke (VIT) are thanked for providing pedigree data, and thanks go to G. von Gadow for providing pictures of cases.
Data Availability
All relevant data are within the paper and its Supporting Information files. Next Generation Sequences were deposited with the European Nucleotide Archive (ENA) under accession number PRJEB20549 (http://www.ebi.uk/ena/data/view/PRJEB20549).
Funding Statement
We acknowledge support by the German Research Foundation and the Open Access Publication Funds of the University of Goettingen. The German Research Foundation and the Open Access Publication Funds of the University of Goettingen only cover the publication costs and did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pascolini D, Mariotti SP. Global estimates of visual impairment: 2010. Br J Ophthalmol. 2012;96(5):614–8. Epub 2011/12/03. doi: 10.1136/bjophthalmol-2011-300539 . [DOI] [PubMed] [Google Scholar]
- 2.Shiels A, Mackay D, Bennett T. Cat-Map [updated 2014/01/10; cited 2016 2016/12/01]. Available from: http://cat-map.wustl.edu.
- 3.Hejtmancik JF. Congenital cataracts and their molecular genetics. Seminars in cell & developmental biology. 2008;19(2):134–49. Epub 2007/11/24. doi: 10.1016/j.semcdb.2007.10.003 ; PubMed Central PMCID: PMC2288487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krump L, O'Grady L, Lorenz I, Grimes T. Congenital cataracts in an Ayrshire herd: a herd case report. Irish veterinary journal. 2014;67(1):2 Epub 2014/01/28. doi: 10.1186/2046-0481-67-2 ; PubMed Central PMCID: PMC3905159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Detlefson JA, Yapp WW. The Inheritance of Congenital Cataract in Cattle. The American Naturalist. 1920;54(632):277–80. doi: 10.2307/2455981 [Google Scholar]
- 6.Carter AH. An inherited blindness (cataract) in cattle. Proceedings of the New Zealand Society of Animal Production. 1960;20:108. [Google Scholar]
- 7.Ashton Barnett KC, Clay CE Clegg FG. Congenital nuclear cataracts in cattle. Vet Rec. 1977;100(24):505–8. Epub 1977/06/11. . [DOI] [PubMed] [Google Scholar]
- 8.Hurley S. Blind but healthy calves. Vet Rec. 2008;163(19):579 . [DOI] [PubMed] [Google Scholar]
- 9.Gregory PW, Mead SW, Regan WM. A congenital hereditary eye defect of cattle. The Journal of heredity. 1943;34:125–8. [Google Scholar]
- 10.Saunders LZ, Fincher MG. Hereditary multiple eye defects in grade Jersey calves. Cornell Vet. 1951;41(4):351–66. Epub 1951/10/01. . [PubMed] [Google Scholar]
- 11.Gelatt KN. Cataracts in cattle. Journal of the American Veterinary Medical Association. 1971;159(2):195–200. Epub 1971/07/15. . [PubMed] [Google Scholar]
- 12.Murgiano L, Jagannathan V, Calderoni V, Joechler M, Gentile A, Drogemuller C. Looking the cow in the eye: deletion in the NID1 gene is associated with recessive inherited cataract in Romagnola cattle. PLoS One. 2014;9(10):e110628 Epub 2014/10/28. doi: 10.1371/journal.pone.0110628 ; PubMed Central PMCID: PMC4210201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graw J. Genetics of crystallins: cataract and beyond. Exp Eye Res. 2009;88(2):173–89. doi: 10.1016/j.exer.2008.10.011 . [DOI] [PubMed] [Google Scholar]
- 14.Graw J. The genetic and molecular basis of congenital eye defects. Nature reviews Genetics. 2003;4(11):876–88. Epub 2003/11/25. doi: 10.1038/nrg1202 . [DOI] [PubMed] [Google Scholar]
- 15.Cheong SS, Hentschel L, Davidson AE, Gerrelli D, Davie R, Rizzo R, et al. Mutations in CPAMD8 Cause a Unique Form of Autosomal-Recessive Anterior Segment Dysgenesis. Am J Hum Genet. 2016;99:1338–1352. doi: 10.1016/j.ajhg.2016.09.022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li ZF, Wu XH, Engvall E. Identification and characterization of CPAMD8, a novel member of the complement 3/alpha2-macroglobulin family with a C-terminal Kazal domain. Genomics. 2004;83(6):1083–93. Epub 2004/06/05. doi: 10.1016/j.ygeno.2003.12.005 . [DOI] [PubMed] [Google Scholar]
- 17.Armstrong PB, Quigley JP. Alpha2-macroglobulin: an evolutionarily conserved arm of the innate immune system. Dev Comp Immunol. 1999;23(4–5):375–90. . [DOI] [PubMed] [Google Scholar]
- 18.Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20(1):34–50. doi: 10.1038/cr.2009.139 . [DOI] [PubMed] [Google Scholar]
- 19.Kenny EE, Pe'er I, Karban A, Ozelius L, Mitchell AA, Ng SM, et al. A genome-wide scan of Ashkenazi Jewish Crohn's disease suggests novel susceptibility loci. PLoS Genet. 2012;8(3):e1002559 Epub 2012/03/14. doi: 10.1371/journal.pgen.1002559 ; PubMed Central PMCID: PMC3297573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18(4):767–78. Epub 2008/11/18. doi: 10.1093/hmg/ddn388 ; PubMed Central PMCID: PMC4334814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sousounis K, Athippozhy AT, Voss SR, Tsonis PA. Plasticity for axolotl lens regeneration is associated with age-related changes in gene expression. Regeneration (Oxf). 2014;1(3):47–57. doi: 10.1002/reg2.25 ; PubMed Central PMCID: PMCPMC4895297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garbe JR, Da Y. Pedigraph: A Software Tool for the Graphing and Analysis of Large Complex Pedigree. User manual Version 2.4. Department of Animal Science, University of Minnesota; 2008. [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. Epub 2004/08/07. doi: 10.1093/bioinformatics/bth457 . [DOI] [PubMed] [Google Scholar]
- 24.Brubaker RF. Flow of aqueous humor in humans [The Friedenwald Lecture]. Invest Ophthalmol Vis Sci. 1991;32(13):3145–66. . [PubMed] [Google Scholar]
- 25.To CH, Kong CW, Chan CY, Shahidullah M, Do CW. The mechanism of aqueous humour formation. Clin Exp Optom. 2002;85(6):335–49. . [PubMed] [Google Scholar]
- 26.Freddo TF, Bartels SP, Barsotti MF, Kamm RD. The source of proteins in the aqueous humor of the normal rabbit. Invest Ophthalmol Vis Sci. 1990;31(1):125–37. . [PubMed] [Google Scholar]
- 27.Goel R, Murthy KR, Srikanth SM, Pinto SM, Bhattacharjee M, Kelkar DS, et al. Characterizing the normal proteome of human ciliary body. Clin Proteomics. 2013;10(1):9 doi: 10.1186/1559-0275-10-9 ; PubMed Central PMCID: PMCPMC3750387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett KL, Funk M, Tschernutter M, Breitwieser FP, Planyavsky M, Ubaida Mohien C, et al. Proteomic analysis of human cataract aqueous humour: Comparison of one-dimensional gel LCMS with two-dimensional LCMS of unlabelled and iTRAQ(R)-labelled specimens. J Proteomics. 2011;74(2):151–66. doi: 10.1016/j.jprot.2010.10.002 . [DOI] [PubMed] [Google Scholar]
- 29.Ji Y, Rong X, Ye H, Zhang K, Lu Y. Proteomic analysis of aqueous humor proteins associated with cataract development. Clin Biochem. 2015;48(18):1304–9. doi: 10.1016/j.clinbiochem.2015.08.006 . [DOI] [PubMed] [Google Scholar]
- 30.Murthy KR, Rajagopalan P, Pinto SM, Advani J, Murthy PR, Goel R, et al. Proteomics of human aqueous humor. OMICS. 2015;19(5):283–93. doi: 10.1089/omi.2015.0029 . [DOI] [PubMed] [Google Scholar]
- 31.Kurien BT, Dorri Y, Dillon S, Dsouza A, Scofield RH. An overview of Western blotting for determining antibody specificities for immunohistochemistry. Methods Mol Biol. 2011;717:55–67. doi: 10.1007/978-1-61779-024-9_3 . [DOI] [PubMed] [Google Scholar]
- 32.De Maria A, Wilmarth PA, David LL, Bassnett S. Proteomic Analysis of the Bovine and Human Ciliary Zonule. Invest Ophthalmol Vis Sci. 2017;58(1):573–85. doi: 10.1167/iovs.16-20866 ; PubMed Central PMCID: PMCPMC5283081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murthy KR, Goel R, Subbannayya Y, Jacob HK, Murthy PR, Manda SS, et al. Proteomic analysis of human vitreous humor. Clin Proteomics. 2014;11(1):29 doi: 10.1186/1559-0275-11-29 ; PubMed Central PMCID: PMCPMC4106660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murthy KR, Dammalli M, Pinto SM, Murthy KB, Nirujogi RS, Madugundu AK, et al. A Comprehensive Proteomics Analysis of the Human Iris Tissue: Ready to Embrace Postgenomics Precision Medicine in Ophthalmology? OMICS. 2016;20(9):510–9. doi: 10.1089/omi.2016.0100 . [DOI] [PubMed] [Google Scholar]
- 35.Zhang P, Kirby D, Dufresne C, Chen Y, Turner R, Ferri S, et al. Defining the proteome of human iris, ciliary body, retinal pigment epithelium, and choroid. Proteomics. 2016;16(7):1146–53. doi: 10.1002/pmic.201500188 ; PubMed Central PMCID: PMCPMC4833206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skeie JM, Mahajan VB. Proteomic landscape of the human choroid-retinal pigment epithelial complex. JAMA Ophthalmol. 2014;132(11):1271–81. doi: 10.1001/jamaophthalmol.2014.2065 . [DOI] [PubMed] [Google Scholar]
- 37.Duckman RH. Visual Development, Diagnosis, and Treatment of the Pediatric Patient: Lippincott Williams & Wilkins; 2006. [Google Scholar]
- 38.Crookston KP, Webb DJ, Wolf BB, Gonias SL. Classification of alpha 2-macroglobulin-cytokine interactions based on affinity of noncovalent association in solution under apparent equilibrium conditions. J Biol Chem. 1994;269(2):1533–40. . [PubMed] [Google Scholar]
- 39.Pelton RW, Saxena B, Jones M, Moses HL, Gold LI. Immunohistochemical localization of TGF beta 1, TGF beta 2, and TGF beta 3 in the mouse embryo: expression patterns suggest multiple roles during embryonic development. J Cell Biol. 1991;115(4):1091–105. ; PubMed Central PMCID: PMCPMC2289937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potts JD, Bassnett S, Beebe DC. Expression of transforming growth factor beta in the embryonic avian lens coincides with the presence of mitochondria. Dev Dyn. 1995;203(3):317–23. doi: 10.1002/aja.1002030304 . [DOI] [PubMed] [Google Scholar]
- 41.Chowdhury UR, Madden BJ, Charlesworth MC, Fautsch MP. Proteome analysis of human aqueous humor. Invest Ophthalmol Vis Sci. 2010;51(10):4921–31. doi: 10.1167/iovs.10-5531 ; PubMed Central PMCID: PMCPMC3066620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Iongh RU, Gordon-Thomson C, Chamberlain CG, Hales AM, McAvoy JW. Tgfbeta receptor expression in lens: implications for differentiation and cataractogenesis. Exp Eye Res. 2001;72(6):649–59. doi: 10.1006/exer.2001.1001 . [DOI] [PubMed] [Google Scholar]
- 43.de Iongh RU, Lovicu FJ, Overbeek PA, Schneider MD, Joya J, Hardeman ED, et al. Requirement for TGFbeta receptor signaling during terminal lens fiber differentiation. Development. 2001;128(20):3995–4010. . [DOI] [PubMed] [Google Scholar]
- 44.Schulz MW, Chamberlain CG, McAvoy JW. Inhibition of transforming growth factor-beta-induced cataractous changes in lens explants by ocular media and alpha 2-macroglobulin. Invest Ophthalmol Vis Sci. 1996;37(8):1509–19. . [PubMed] [Google Scholar]
- 45.Lovicu FJ, Schulz MW, Hales AM, Vincent LN, Overbeek PA, Chamberlain CG, et al. TGFbeta induces morphological and molecular changes similar to human anterior subcapsular cataract. Br J Ophthalmol. 2002;86(2):220–6. Epub 2002/01/30. ; PubMed Central PMCID: PMCPmc1771017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cogan DG. Anatomy of lens and pathology of cataracts. Exp Eye Res. 1962;1:291–5. . [DOI] [PubMed] [Google Scholar]
- 47.Bistner SI, Rubin LF, Saunders LZ. The Ocular Lesions of Bovine Viral Diarrhea-Mucosal Disease. Path Vet. 1970;7:275–86. [DOI] [PubMed] [Google Scholar]
- 48.VIT, w.V. VIT. Relative Breeding Value (ranked by RZG), daughter proven, only active AI bulls, German Red Holsteins 2015 [2015/10/01]. Available from: http://www.vit.de/fileadmin/user_upload/vit-fuers-rind/zuchtwertschaetzung/milchrinder-zws-online/RotBunt_RZG_aktiveBullen.pdf.
- 49.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215 Epub 1988/02/11. ; PubMed Central PMCID: PMC334765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–9. doi: 10.1006/abio.1987.9999 . [DOI] [PubMed] [Google Scholar]
- 51.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. Epub 2007/08/19. doi: 10.1086/519795 ; PubMed Central PMCID: PMCPmc1950838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324 ; PubMed Central PMCID: PMCPMC2705234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110 ; PubMed Central PMCID: PMCPMC2928508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. doi: 10.4161/fly.19695 ; PubMed Central PMCID: PMCPMC3679285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Förster T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Annalen der Physik. 1948;437(1–2):55–75. doi: 10.1002/andp.19484370105 [Google Scholar]
- 56.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134 Epub 2012/06/20. doi: 10.1186/1471-2105-13-134 ; PubMed Central PMCID: PMC3412702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schutz E, von Ahsen N. Spreadsheet software for thermodynamic melting point prediction of oligonucleotide hybridization with and without mismatches. BioTechniques. 1999;27(6):1218–22, 24. Epub 2000/01/13. . [DOI] [PubMed] [Google Scholar]
- 58.von Ahsen N, Oellerich M, Armstrong VW, Schutz E. Application of a thermodynamic nearest-neighbor model to estimate nucleic acid stability and optimize probe design: prediction of melting points of multiple mutations of apolipoprotein B-3500 and factor V with a hybridization probe genotyping assay on the LightCycler. Clinical chemistry. 1999;45(12):2094–101. Epub 1999/12/10. . [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein extracts of a normal adult lens (lane 2), normal fetal lens (lane 3) and adult cataractous lens (lane 4) were loaded on a 4–12% gradient SDS-PAGE. As positive control human CPAMD8 over-expression lysate was used (lane 1). Proteins were transferred to nitrocellulose membranes and filters were blocked with 5% non-fat dry milk in TBS-T followed by incubation with anti-CPAMD8 antibody HPA031328 (S2 Table, dilution 1:200). Detection of chemiluminescent signals was performed using Amersham ECL detection reagent and membranes were exposed to X-ray films. CPAMD8 bands in lane 1 (human control lysate), 2 (bovine normal adult lens) and 3 (bovine normal fetal lens) are indicated with an arrow. Protein sizes are indicated in kDa. Scans of X-ray films were cropped using GIMP 2.8.18 (GNU Image Manipulation Program).
(TIFF)
Five μl of in vitro produced bovine CPAMD8 epitopes were loaded on a 4–12% gradient SDS-PAGE (lane 1, lane 2). Proteins were transferred to nitrocellulose membranes and filters were blocked with 5% non-fat dry milk in TBS-T followed by incubation with anti-CPAMD8 antibody HPA031328 (S2 Table, dilution 1:200). Detection of chemiluminescent signals was performed using Amersham ECL detection reagent and membranes were exposed to X-ray films. The band in lane 1 shows the expected size of the epitope (approx. 33 kDa) and is indicated with an arrow. In lane 2 another in vitro expressed epitope lysate was loaded showing no reactivity. Lane 3 shows a negative control lysate. Protein sizes are indicated in kDa. Scans of X-ray films were cropped using GIMP 2.8.18 (GNU Image Manipulation Program).
(TIFF)
Consciousness, posture, gait, swallowing and tongue tone were normal in all cases. Behavior differed from calm alertness to anxiety. If not stated otherwise the ocular fundus was without pathological findings.
(DOCX)
1) Sigma-Aldrich Chemie (Taufkirchen, Germany); 2) OriGene Technologies (Rockville, USA).
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Next Generation Sequences were deposited with the European Nucleotide Archive (ENA) under accession number PRJEB20549 (http://www.ebi.uk/ena/data/view/PRJEB20549).