Abstract
Background
Genome-wide association studies have identified ~100 common genetic variants associated with breast cancer risk, the majority of which were discovered in women of European ancestry. Due to different patterns of linkage disequilibrium, many of these genetic markers may not represent signals in populations of African ancestry.
Methods
We tested 74 breast cancer risk variants and conducted fine-mapping of these susceptibility regions in 6,522 breast cancer cases and 7,643 controls of African ancestry from three genetic consortia (AABC, AMBER and ROOT).
Results
Fifty-four of the 74 variants (73%) were found to have odds ratios that were directionally consistent with those previously reported, of which twelve were nominally statistically significant (P < 0.05). Through fine-mapping, in six regions (3p24, 12p11, 14q13, 16q12/FTO, 16q23, 19p13) we observed seven markers that better represent the underlying risk variant for overall breast cancer or breast cancer subtypes, whereas in another two regions (11q13, 16q12/TOX3) we identified suggestive evidence of signals that are independent of the reported index variant. Overlapping chromatin features and regulatory elements suggest that many of the risk alleles lie in regions with biological functionality.
Conclusions
Through fine-mapping of known susceptibility regions we have revealed alleles that better characterize breast cancer risk in women of African ancestry.
Impact
The risk alleles identified represent genetic markers for modeling and stratifying breast cancer risk in women of African ancestry.
Keywords: Breast cancer, African ancestry, Genome-wide association studies, Fine-mapping
Introduction
Genome-wide association (GWAS) and large-scale fine-mapping studies have led to the identification of >100 breast cancer susceptibility loci that are estimated to explain approximately 20% of the two-fold familial risk of breast cancer in women of European descendant (1–13). For populations of African ancestry, where the span of linkage disequilibrium (LD) has been shortened by recombination events (more generations), a weaker correlation between an ‘index’ marker (from GWAS in Asian and European ancestry populations) and biologically relevant risk variants is expected. As a consequence, the index marker might not accurately capture the risk associated with the biologically functional variant in African ancestry populations. Comprehensive testing in a large African ancestry sample is needed to identify a set of markers that better capture risk associated with the functional allele at known risk regions, which is an important prerequisite for constructing genetic risk models for this population. Previous fine-mapping investigations in women of African ancestry have been limited in size, with the largest study including 3,016 breast cancer cases and 2,745 controls and having 80% power to detect reported effect sizes for only 10 of 72 variants examined (1–15).
To obtain greater statistical power for fine-mapping of known breast cancer susceptibility regions we combined genotype and imputed data for 6,522 breast cancer cases and 7,643 controls from three large consortia of African ancestry breast cancer - the African American Breast Cancer GWAS Consortium (AABC) (16), the African American Breast Cancer Epidemiology and Risk Consortium (AMBER) (17, 18), and the Genome-Wide Association Study of Breast Cancer in the African Diaspora Consortium (ROOT) (19). In addition to testing the reported index variants from previous GWAS, we conducted association analyses and functional annotation across each region in search of markers that might best define breast cancer risk in women of African ancestry.
Materials and Methods
Studies
The genetic data included in this analysis were from three consortia of breast cancer in women of African ancestry (AABC, AMBER and ROOT). For this analysis, the African American Breast Cancer Consortium (AABC) included seven epidemiologic studies: The Multiethnic Cohort study (MEC), 734/1,003; The Los Angeles component of The Women’s Contraceptive and Reproductive Experiences (CARE) Study, 380/224; The San Francisco Bay Area Breast Cancer Study (SFBCS), 172/23; The Northern California site of the Breast Cancer Family Registry (NC-BCFR), 440/53; The Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) Cohort, 64/133; The Nashville Breast Health Study (NBHS), 310/186; and The Wake Forest University Breast Cancer Study (WFBC), 125/153). The present analysis includes GWAS data for 2,225 invasive cases and 1,983 controls from AABC (14). Although the Women’s Circle of Health Study (WCHS) and The Carolina Breast Cancer Study (CBCS) participated in AABC, samples from those studies are included as part of the AMBER consortium described below.
The AMBER consortium (18) included three studies for a total of 2,754 invasive breast cancer cases and 3,698 controls: the Black Women’s Health Study (BWHS) (20) (752/2249); WCHS (681/834) (21); and CBCS (1321/615) (22).
The ROOT consortium (19) included six studies and a total of 1,657 cases and 2,028 controls of African ancestry: The Nigerian Breast Cancer Study (NBCS), 711/623; The Barbados National Cancer Study (BNCS), 92/229; The Racial Variability in Genotypic Determinants of Breast Cancer Risk Study (RVGBC), 145/257; The Baltimore Breast Cancer Study (BBCS), 95/102; The Chicago Cancer Prone Study (CCPS), 394/387; and The Southern Community Cohort (SCCS), 220/430.
Genotyping and Quality Control
Genotyping in AABC was conducted using the Illumina Human 1M-Duo BeadChip as described in Chen et al. (14). The ROOT samples were genotyped using the Illumina 2.5M array (19). Samples in AMBER were genotyped using an Illumina Infinium custom ~160K SNP array which included ~45,000 SNPs selected primarily for fine-mapping of known breast cancer susceptibility regions.
Statistical Analysis
Imputation in AABC and AMBER was conducted using IMPUTE2 (23) to a cosmopolitan panel of all 1000 Genome Project subjects (March 2012 release). MACH v1.0 was used to impute the unobserved SNPs in ROOT in YRI and CEU haplotype data from HapMap Phased II (Release #22) and phased African haplotype data from the 1000 Genome Project (November 2010 release) (24). Imputed SNPs with imputation quality score > 0.7 and a minor allele frequency > 0.01 in each study were used in the fine-mapping analysis. We examined 74 risk variants for breast cancer in 72 regions that had been reported at the time this study was initiated (1–11). One additional variant, rs11571833 at chromosome 13q13, was not genotyped and could not be imputed in all 3 studies; this variant had a minor allele frequency of 0.006 in the 1000 Genomes AFR population. These 74 risk variants include stronger markers than the index SNP found in GWAS as well as independent signals discovered through subsequent fine-mapping studies (Supplemental Table S1) (1, 3, 4, 8, 10, 11).
A total of 6,522 breast cancer cases (2,933 ER+ and 1,876 ER−) and 7,643 controls were included in the analysis. For each typed and imputed SNP, odds ratios (OR) and 95% confidence intervals (95% CI) were estimated using unconditional logistic regression adjusting for age (at diagnosis for cases and age at the reference date for controls), study, and the first 10 eigenvectors from a principal components analysis (25). For each SNP that existed in all three studies, we tested for allele dosage effects separately in each of the three studies, applying a 1-degree-of-freedom Wald chi-square trend test. Results were then combined using inverse variance-weighted fixed-effects meta-analysis, as implemented in METAL (26). We tested for effect heterogeneity between studies using Cochran’s Q-test as implemented in METAL. Power calculations were conducted using Quanto (http://hydra.usc.edu/gxe/) using the OR in previous GWAS and the allele frequency in African Americans.
To identify alleles that might capture the biologically functional variant at 70 of the known breast cancer risk regions, we searched and tested LD proxies among the genotyped and imputed SNPs that were correlated (r2 ≥ 0.4) with the index SNP [within 250kb or larger if the index signal was contained within an LD block (based on the D′ statistic) of > 250kb] in European ancestry populations, resulting in a total of 157,920 SNPs included in the analysis. Two regions, 5p15 and 20q11, were excluded from fine-mapping because the AABC sample was involved in the discovery of the risk loci in these regions (27, 28). The GWAS arrays and imputation in AAPC, AMBER and ROOT provided good coverage of common variation (>5%) in the fine-mapped regions in African ancestry populations. For AABC, an average of 96% of common SNPs with a MAF >5% in the Phase 3 1000’s Genome AFR population were tagged (at r2>0.8) by the genotyped and imputed SNPs. For ROOT and AMBER, these averages were each 97%. For each study, the coverage was >90% for all regions, with the exception of chromosome 1p11 (45% in each) and chromosome 6q25 in AMBER (82%).
Locus-specific significance levels were calculated as described in Feng et al, 2014 (15) (Supplementary Table S2). More specifically, locus-specific significance levels were calculated as 0.05 divided by the number of tag SNPs in the African population (1000 Genomes, AFR, March 2012 Release) that capture (r2 ≥ 0.8) all SNPs correlated with the index signal in the European population (1000 Genomes, EUR, March 2012 Release). To reduce false-positive signals, we required the P-value of all better markers to be less than 0.01. In an attempt to eliminate minor fluctuations in P-values for correlated SNPs, we also required the P-value to decrease by more than one order of magnitude compared with the association with the index signal. If multiple variants satisfied the above criterion in each region, only the most statistically significant variant was reported.
We also tested for novel independent associations, focusing on all genotyped and imputed SNPs in each region that were uncorrelated with the index signal in European ancestry populations (r2 < 0.1), and applied a significance criterion of α = 5×10−6 for defining suggestive novel associations, as used in prior studies. (14) This α is not as conservative as genome-wide significance and is an approximation of the number of tests to capture (at r2 ≥ 0.8) common risk alleles across all regions. In order to confirm independent associations, conditional analyses were performed that included the index SNP or better marker plus the most significant uncorrelated allele. The analysis was first conducted in each separate study and then combined using fixed-effects meta-analysis. Haplotype analysis on 16q12 was conducted applying the “haplo.stats” package in R (http://www.mayo.edu/research/labs/statistical-genetics-genetic-epidemiology/software).
The procedures described above were applied to the analysis of overall breast cancer as well as in secondary analyses stratified by ER status.
Functional Annotations
We assessed whether any of the signals co-localized with 65 chromatin features that capture open chromatin regions and regulatory elements across the genome in ER+ breast cancer (MCF7, T47D, HCC1954), ER− breast cancer (MDAMB231) and normal breast (HMEC, Myoepithelial, Fibroblast, Luminal epithelial) cells identified by the Coetzee Laboratory (29–31) or obtained from the Encyclopedia of DNA Elements (ENCODE) project (32) or NIH Roadmap Epigenomics Mapping Consortium (REMC) (33, 34). Enriched regions of chromatin features were either called by using the Sole-search program (35) or obtained from GEO databases (GSE35583, GSE32970, GSE35239, GSE46074, GSE49651, GSE78913). To refine the genomic regulatory regions, chromatin state segmentation information built by using a Hidden Markov Model (HMM) in MCF7 and HMEC were also included (36, 37).
We used motifbreakR (38) to search for transcription factor motifs that bind to each variant (39–43). Chromatin features that overlapped variants and motifs that significantly altered binding (using the default setting with the score threshold, 0.9) are summarized in Supplementary Table S3. We also included key transcription factors for breast cancer such as FOXA1, GATA3, and ESR1 ChIP-seq data in ER+ breast cancer cells (MCF7, T47D) from the ENCODE project to examine the occupancy of transcription factors in vitro at regulatory elements where variants reside (32).
Results
Of the 74 breast cancer risk variants, 68 were also common in women of African ancestry, with minor allele frequencies greater than 0.05 in all three studies. Of these 68 variants, we had ≥ 50% and ≥ 80% power (at p<0.05) to detect previously reported effect sizes for 51 and 36 variants, respectively. The odds ratios observed for 54 (73%) of the 74 SNPs were directionally consistent with those previously reported (i.e. ORs were in the same direction), with 12 variants nominally statistically significant at P < 0.05 (Table 1; Supplementary Table S1). Of the 61 SNPs that were not replicated at P < 0.05 in this study, statistical power to detect the previously reported effect size for overall breast cancer was ≥ 80% for 29 (48%) SNPs (Supplementary Table S1). Fifty-three (72%) variants were positively associated with ER+ breast cancer (8 statistically significant at P < 0.05) and 37 (50%) variants were positively associated with ER− disease (10 statistically significant at P < 0.05) (Supplementary Table S4). Of the 7 variants that were reported to be more strongly associated with ER− than ER+ disease in European ancestry populations (rs6678914/1q32, rs4245739/1q32, rs12710696/2p24, rs10069690/5p15, rs11075995/16q12, rs67397200/19p13, rs2284378/20q11) (2, 27, 28, 44), all were positively associated with risk of ER− disease (3 at P < 0.05; rs4245739, rs10069690, and rs67397200). Statistical power was ≥ 80% to detect the reported effect size with ER− disease for 4 of the 7 variants (Supplementary Table S4).
Table 1.
Associations of 74 Breast Cancer Risk Variants with Risk of Overall Breast Cancer and Breast Cancer Subtype in Women of African Ancestry.
| Initial GWAS | META Analysis Results | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPID | Chr | Positiona | Allelesb | Nearest Gene | RAFc | OR (95% CI) | P | RAFc | OR (95% CI) | P | ER+ OR (95% CI) | ER+ P | ER− OR (95% CI) | ER− P | Phet |
| rs616488 | 1 | 10566215 | A/G | PEX14 | 0.67 | 1.06(1.04–1.09) | 2.0×10−10 | 0.86 | 1.07(0.99–1.16) | 0.080 | 1.04 (0.94–1.15) | 0.43 | 1.11 (0.98–1.25) | 0.10 | 0.29 |
| rs12022378 | 1 | 114448389 | A/G | PTPN22:BCL2L15: AP4B1:DCLRE1B: HIPK1 | 0.17 | 1.07(1.04–1.09) | 1.8×10−8 | 0.047 | 0.99(0.87–1.14) | 0.94 | 0.98 (0.83–1.16) | 0.80 | 1.00 (0.81–1.22) | 0.96 | 0.85 |
| rs11249433 | 1 | 121280613 | G/A | NA | 0.41 | 1.09(1.07–1.11) | 2.0×10−26 | 0.14 | 1.03(0.95–1.11) | 0.51 | 1.03 (0.93–1.14) | 0.52 | 1.02 (0.91–1.15) | 0.73 | 0.21 |
| rs6678914 | 1 | 202187176 | G/A | LGR6 | 0.59 | 1.00(0.98–1.02) | 0.86 | 0.66 | 1.01(0.95–1.07) | 0.68 | 0.98 (0.91–1.05) | 0.54 | 1.06 (0.97–1.15) | 0.19 | 0.92 |
| rs4245739 | 1 | 204518842 | C/A | MDM4 | 0.26 | 1.02(1.00–1.04) | 0.080 | 0.24 | 1.02(0.97–1.09) | 0.41 | 0.96 (0.89–1.03) | 0.27 | 1.14 (1.05–1.25) | 3.0×10−3 | 0.030 |
| rs12710696 | 2 | 19320803 | A/G | NA | 0.36 | 1.04(1.01–1.06) | 9.7×10−4 | 0.52 | 1.05(1.00–1.10) | 0.048 | 1.05 (0.98–1.12) | 0.13 | 1.07 (0.99–1.15) | 0.11 | 0.37 |
| rs4849887 | 2 | 121245122 | G/A | NA | 0.90 | 1.10(1.06–1.14) | 3.7×10−11 | 0.71 | 1.09(1.03–1.16) | 1.4×10−3 | 1.11 (1.03–1.19) | 4.9×10−3 | 1.14 (1.05–1.24) | 2.5×10−3 | 0.46 |
| rs2016394 | 2 | 172972971 | G/A | METAP1D:DLX1: DLX2 | 0.52 | 1.05(1.03–1.08) | 1.2×10−8 | 0.72 | 1.02(0.96–1.08) | 0.58 | 1.04 (0.96–1.12) | 0.32 | 1.01 (0.92–1.10) | 0.83 | 0.75 |
| rs1550623 | 2 | 174212894 | A/G | CDCA7 | 0.84 | 1.06(1.03–1.09) | 3.0×10−8 | 0.70 | 1.02(0.97–1.08) | 0.37 | 1.06 (0.99–1.14) | 0.11 | 1.02 (0.94–1.11) | 0.65 | 0.047 |
| rs1830298e | 2 | 202181247 | C/T | CASP8/ALS2CR12 | 0.29 | 1.05(1.03–1.07) | 1.0×10−5 | 0.23 | 1.02(0.96–1.09) | 0.43 | 0.96 (0.88–1.04) | 0.28 | 0.98 (0.89–1.07) | 0.63 | 0.98 |
| rs4442975e | 2 | 217920769 | G/T | IGFBP5 | 0.5 | 1.15(1.12–1.16) | 3.9×10−46 | 0.63 | 1.07(1.02–1.13) | 8.2×10−3 | 1.13 (1.06–1.21) | 5.1×10−4 | 1.01 (0.93–1.10) | 0.74 | 0.11 |
| rs34005590e | 2 | 217963060 | C/A | IGFBP5 | 0.95 | 1.22(1.16–1.28) | 5.6×10−17 | 0.89 | 1.08(0.99–1.17) | 0.083 | 0.87 (0.77–0.97) | 0.014 | 1.06 (0.93–1.21) | 0.40 | 0.38 |
| rs16857609 | 2 | 218296508 | A/G | DIRC3 | 0.26 | 1.08(1.06–1.10) | 1.1×10−15 | 0.24 | 1.13(1.07–1.20) | 3.2×10−5 | 1.15 (1.07–1.24) | 2.1×10−4 | 1.15 (1.05–1.25) | 2.6×10−3 | 0.49 |
| rs6762644 | 3 | 4742276 | G/A | ITPR1:EGOT | 0.40 | 1.07(1.04–1.09) | 2.2×10−12 | 0.46 | 1.01(0.96–1.07) | 0.63 | 1.05 (0.98–1.12) | 0.18 | 0.96 (0.89–1.03) | 0.27 | 0.39 |
| rs4973768 | 3 | 27416013 | A/G | SLC4A7 | 0.48 | 1.10(1.08–1.12) | 2.3×10−30 | 0.36 | 1.01(0.96–1.06) | 0.76 | 1.02 (0.95–1.09) | 0.59 | 0.96 (0.88–1.04) | 0.31 | 0.35 |
| rs12493607 | 3 | 30682939 | G/C | TGFBR2 | 0.35 | 1.06(1.03–1.08) | 2.3×10−8 | 0.14 | 1.03(0.96–1.11) | 0.40 | 1.06 (0.96–1.17) | 0.24 | 0.99 (0.88–1.11) | 0.87 | 0.85 |
| rs9790517 | 4 | 106084778 | A/G | TET2 | 0.23 | 1.05(1.03–1.08) | 4.2×10−8 | 0.09 | 0.94(0.85–1.04) | 0.22 | 0.88 (0.77–1.00) | 0.051 | 0.94 (0.80–1.10) | 0.42 | 0.082 |
| rs6828523 | 4 | 175846426 | C/A | ADAM29 | 0.87 | 1.11(1.09–1.15) | 3.5×10−16 | 0.65 | 1.05(0.99–1.10) | 0.096 | 1.06 (0.99–1.13) | 0.099 | 1.05 (0.97–1.14) | 0.21 | 0.97 |
| rs10069690 | 5 | 1279790 | A/G | TERT | 0.27 | 1.06(1.04–1.09) | 7.2×10−9 | 0.56 | 1.12(1.07–1.18) | 8.4×10−6 | 1.06 (0.99–1.13) | 0.097 | 1.31 (1.21–1.42) | 5.5×10−11 | 0.88 |
| rs10941679 | 5 | 44706498 | G/A | NA | 0.27 | 1.13(1.10–1.15) | 1.7×10−37 | 0.21 | 1.02(0.96–1.09) | 0.52 | 1.08 (1.00–1.17) | 0.059 | 0.93 (0.84–1.02) | 0.14 | 0.65 |
| rs62355902e | 5 | 56053723 | T/A | MAP3K1 | 0.16 | 1.21(1.19–1.24) | 9.5×10−49 | 0.10 | 1.08(0.99–1.18) | 0.070 | 1.18 (1.06–1.31) | 3.2×10−3 | 0.95 (0.83–1.09) | 0.46 | 0.64 |
| rs10472076 | 5 | 58184061 | G/A | RAB3C | 0.38 | 1.05(1.03–1.07) | 2.9×10−8 | 0.28 | 0.96(0.91–1.02) | 0.16 | 0.95 (0.89–1.03) | 0.21 | 0.98 (0.90–1.07) | 0.64 | 0.81 |
| rs1353747 | 5 | 58337481 | A/C | PDE4D | 0.91 | 1.09(1.05–1.12) | 2.5×10−8 | 0.98 | 0.93(0.77–1.13) | 0.45 | 0.89 (0.71–1.13) | 0.36 | 0.82 (0.61–1.09) | 0.17 | 0.82 |
| rs1432679 | 5 | 158244083 | G/A | EBF1 | 0.43 | 1.07(1.05–1.09) | 2.0×10−14 | 0.79 | 1.09(1.02–1.16) | 0.011 | 1.04 (0.95–1.13) | 0.41 | 1.22 (1.10–1.35) | 1.2×10−4 | 0.065 |
| rs11242675 | 6 | 1318878 | A/G | FOXQ1 | 0.61 | 1.06(1.04–1.09) | 7.1×10−9 | 0.52 | 1.01(0.96–1.06) | 0.82 | 1.01 (0.94–1.07) | 0.88 | 1.04 (0.96–1.12) | 0.35 | 0.19 |
| rs204247 | 6 | 13722523 | G/A | RANBP9 | 0.43 | 1.05(1.03–1.07) | 8.3×10−9 | 0.34 | 1.06(1.01–1.12) | 0.024 | 1.07 (1.00–1.14) | 0.064 | 1.05 (0.97–1.14) | 0.21 | 0.069 |
| rs17529111 | 6 | 82128386 | G/A | NA | 0.22 | 1.06(1.04–1.09) | 4.3×10−9 | 0.08 | 0.98(0.88–1.08) | 0.68 | 1.00 (0.88–1.14) | 0.99 | 0.96 (0.82–1.12) | 0.59 | 0.070 |
| rs9485372 | 6 | 149608874 | G/A | TAB2 | 0.55 | 1.11(1.09–1.15) | 3.8×10−12 | 0.80 | 1.02(0.96–1.09) | 0.50 | 1.04 (0.95–1.14) | 0.37 | 0.98 (0.88–1.08) | 0.63 | 0.81 |
| rs3757322e | 6 | 151942194 | G/T | ESR1 | 0.33 | 1.09(1.07–1.11) | 8.1×10−17 | 0.48 | 1.04(0.99–1.09) | 0.16 | 1.00 (0.93–1.06) | 0.89 | 1.08 (1.00–1.17) | 0.054 | 0.023 |
| rs9397437e | 6 | 151952332 | A/G | ESR1 | 0.07 | 1.20(1.16–1.25) | 4.0×10−22 | 0.047 | 1.08(0.96–1.22) | 0.19 | 1.02 (0.87–1.19) | 0.82 | 1.16 (0.97–1.38) | 0.097 | 0.99 |
| rs720475 | 7 | 144074929 | G/A | ARHGEF5:NOBOX | 0.75 | 1.06(1.04–1.09) | 7.0×10−11 | 0.88 | 1.00(0.92–1.08) | 0.98 | 1.03 (0.93–1.14) | 0.58 | 1.00 (0.88–1.13) | 0.95 | 0.52 |
| rs9693444 | 8 | 29509616 | A/C | NA | 0.32 | 1.07(1.05–1.09) | 9.2×10−14 | 0.37 | 1.04(0.99–1.10) | 0.11 | 1.03 (0.97–1.11) | 0.33 | 1.04 (0.96–1.12) | 0.36 | 0.88 |
| rs6472903 | 8 | 76230301 | A/C | NA | 0.82 | 1.10(1.08–1.12) | 1.7×10−17 | 0.90 | 1.07(0.98–1.16) | 0.13 | 1.05 (0.94–1.17) | 0.40 | 1.01 (0.88–1.14) | 0.93 | 0.86 |
| rs2943559 | 8 | 76417937 | G/A | HNF4G | 0.070 | 1.13(1.09–1.17) | 5.7×10−15 | 0.21 | 1.06(1.00–1.12) | 0.071 | 1.03 (0.95–1.12) | 0.46 | 1.11 (1.01–1.21) | 0.033 | 0.63 |
| rs13281615 | 8 | 128355618 | G/A | NA | 0.42 | 1.09(1.07–1.12) | 9.6×10−28 | 0.43 | 1.03(0.98–1.08) | 0.21 | 1.04 (0.97–1.11) | 0.24 | 0.99 (0.92–1.07) | 0.84 | 0.56 |
| rs11780156 | 8 | 129194641 | A/G | MIR1208 | 0.16 | 1.07(1.04–1.10) | 3.4×10−11 | 0.05 | 0.91(0.80–1.03) | 0.14 | 0.90 (0.76–1.06) | 0.21 | 0.98 (0.81–1.20) | 0.86 | 0.60 |
| rs1011970 | 9 | 22062134 | A/C | CDKN2A/B | 0.17 | 1.06(1.03–1.08) | 5.5×10−8 | 0.31 | 1.04(0.99–1.10) | 0.15 | 1.07 (1.00–1.15) | 0.055 | 0.97 (0.90–1.06) | 0.50 | 0.37 |
| rs10759243 | 9 | 110306115 | A/C | NA | 0.39 | 1.06(1.03–1.08) | 1.2×10−8 | 0.58 | 0.99(0.94–1.04) | 0.75 | 1.02 (0.96–1.10) | 0.49 | 0.96 (0.88–1.04) | 0.28 | 0.71 |
| rs13294895e | 9 | 110837176 | T/C | NA | 0.18 | 1.09(1.06–1.12) | 3.0×10−11 | 0.048 | 1.02(0.89–1.17) | 0.76 | 1.06 (0.89–1.25) | 0.53 | 1.00 (0.81–1.23) | 0.99 | 0.63 |
| rs676256e | 9 | 110895353 | T/C | NA | 0.62 | 1.11(1.09–1.13) | 1.6×10−25 | 0.75 | 1.07(1.01–1.14) | 0.018 | 1.08 (1.00–1.16) | 0.061 | 1.05 (0.96–1.15) | 0.26 | 0.39 |
| rs2380205 | 10 | 5886734 | G/A | ANKRD16 | 0.56 | 1.02(1.00–1.04) | 2.1×10−3 | 0.42 | 0.98(0.94–1.04) | 0.54 | 0.95 (0.89–1.02) | 0.16 | 0.99 (0.91–1.07) | 0.75 | 0.99 |
| rs7072776 | 10 | 22032942 | A/G | MLLT10:DNAJC1 | 0.29 | 1.07(1.05–1.09) | 4.3×10−14 | 0.50 | 1.03(0.98–1.09) | 0.19 | 1.02 (0.96–1.09) | 0.48 | 1.07 (0.99–1.16) | 0.079 | 0.59 |
| rs11814448 | 10 | 22315843 | C/A | DNAJC1 | 0.020 | 1.26(1.18–1.35) | 9.3×10−16 | 0.59 | 1.04(0.98–1.10) | 0.16 | 1.03 (0.96–1.11) | 0.43 | 1.09 (1.00–1.19) | 0.047 | 0.54 |
| rs10995190 | 10 | 64278682 | G/A | ZNF365 | 0.85 | 1.16(1.14–1.19) | 1.3×10−36 | 0.84 | 0.96(0.90–1.03) | 0.28 | 0.99 (0.91–1.08) | 0.84 | 0.94 (0.85–1.04) | 0.26 | 0.80 |
| rs704010 | 10 | 80841148 | A/G | ZMIZ1 | 0.39 | 1.08(1.06–1.10) | 7.4×10−22 | 0.11 | 1.10(1.00–1.21) | 0.043 | 1.07 (0.95–1.20) | 0.26 | 1.08 (0.94–1.25) | 0.27 | 0.78 |
| rs7904519 | 10 | 114773927 | G/A | TCF7L2 | 0.46 | 1.06(1.04–1.08) | 3.1×10−8 | 0.78 | 1.06(0.99–1.13) | 0.087 | 1.07 (0.98–1.16) | 0.12 | 1.11 (1.01–1.23) | 0.038 | 0.11 |
| rs11199914 | 10 | 123093901 | G/A | NA | 0.68 | 1.05(1.04–1.08) | 1.9×10−8 | 0.49 | 1.02(0.96–1.07) | 0.53 | 1.03 (0.97–1.11) | 0.34 | 0.93 (0.86–1.01) | 0.069 | 0.66 |
| rs2981579 | 10 | 123337335 | A/G | FGFR2 | 0.43 | 1.27(1.24–1.29) | 1.9×10−170 | 0.59 | 1.14(1.08–1.21) | 7.2×10−6 | 1.16 (1.09–1.24) | 9.9×10−6 | 1.09 (1.01–1.18) | 0.027 | 0.84 |
| rs3817198 | 11 | 1909006 | G/A | LSP1 | 0.32 | 1.07(1.05–1.09) | 1.5×10−11 | 0.17 | 0.99(0.92–1.06) | 0.78 | 1.01 (0.92–1.10) | 0.83 | 0.94 (0.85–1.05) | 0.29 | 2.8×10−3 |
| rs3903072 | 11 | 65583066 | C/A | DKFZp761E198: OVOL1:SNX32:CFL1: MUS81 | 0.53 | 1.05(1.04–1.08) | 8.6×10−12 | 0.81 | 0.98(0.92–1.06) | 0.67 | 0.99 (0.91–1.09) | 0.91 | 0.93 (0.84–1.04) | 0.21 | 0.28 |
| rs614367 | 11 | 69328764 | A/G | NA | 0.16 | 1.21(1.18–1.24) | 2.2×10−63 | 0.13 | 1.03(0.96–1.11) | 0.46 | 1.01 (0.92–1.11) | 0.84 | 0.95 (0.85–1.07) | 0.42 | 0.30 |
| rs11820646 | 11 | 129461171 | G/A | NA | 0.59 | 1.05(1.03–1.08) | 1.1×10–9 | 0.77 | 0.97(0.91–1.04) | 0.46 | 0.93 (0.86–1.01) | 0.088 | 0.97 (0.88–1.06) | 0.47 | 0.62 |
| rs12422552 | 12 | 14413931 | C/G | NA | 0.26 | 1.05(1.03–1.07) | 3.7×10−8 | 0.41 | 1.01(0.96–1.07) | 0.65 | 1.03 (0.97–1.11) | 0.33 | 1.00 (0.92–1.08) | 0.90 | 0.95 |
| rs7297051e | 12 | 28174817 | C/T | NA | 0.76 | 1.14(1.11–1.16) | 4.0×10−28 | 0.88 | 1.05(0.97–1.13) | 0.24 | 1.08 (0.98–1.19) | 0.14 | 1.06 (0.94–1.19) | 0.35 | 0.90 |
| rs17356907 | 12 | 96027759 | A/G | NTN4 | 0.70 | 1.10(1.08–1.12) | 1.8×10−22 | 0.79 | 1.06(1.00–1.13) | 0.057 | 1.10 (1.01–1.20) | 0.022 | 1.01 (0.92–1.11) | 0.81 | 0.21 |
| rs1292011 | 12 | 115836522 | A/G | NA | 0.59 | 1.09(1.06–1.11) | 8.9×10−22 | 0.55 | 0.98(0.94–1.03) | 0.52 | 1.01 (0.94–1.07) | 0.85 | 0.93 (0.86–1.00) | 0.052 | 0.31 |
| rs2236007 | 14 | 37132769 | G/A | PAX9:SLC25A21 | 0.79 | 1.08(1.05–1.10) | 1.7×10−13 | 0.92 | 1.00(0.91–1.11) | 0.93 | 0.98 (0.86–1.11) | 0.72 | 0.94 (0.80–1.09) | 0.39 | 0.094 |
| rs2588809 | 14 | 68660428 | A/G | RAD51L1 | 0.16 | 1.08(1.05–1.11) | 1.4×10−10 | 0.28 | 1.00(0.95–1.06) | 0.88 | 1.06 (0.98–1.14) | 0.12 | 0.96 (0.89–1.05) | 0.40 | 0.73 |
| rs999737 | 14 | 69034682 | G/A | RAD51L1 | 0.78 | 1.09(1.06–1.11) | 2.5×10−19 | 0.94 | 1.05(0.92–1.19) | 0.49 | 1.06 (0.90–1.24) | 0.52 | 1.03 (0.84–1.26) | 0.80 | 0.37 |
| rs941764 | 14 | 91841069 | G/A | CCDC88C | 0.34 | 1.06(1.04–1.09) | 3.7×10−10 | 0.69 | 1.02(0.96–1.08) | 0.52 | 1.01 (0.94–1.09) | 0.77 | 1.02 (0.93–1.11) | 0.67 | 0.017 |
| rs3803662 | 16 | 52586341 | A/G | TOX3 | 0.29 | 1.24(1.21–1.27) | 2.1×10−114 | 0.50 | 1.00(0.95–1.05) | 0.89 | 1.01 (0.95–1.08) | 0.73 | 0.98 (0.91–1.06) | 0.67 | 0.091 |
| rs17817449 | 16 | 53813367 | A/C | MIR1972-2:FTO | 0.60 | 1.08(1.05–1.10) | 6.4×10−14 | 0.61 | 1.07(1.01–1.12) | 0.012 | 1.08 (1.01–1.15) | 0.029 | 1.04 (0.96–1.13) | 0.32 | 0.86 |
| rs11075995 | 16 | 53855291 | A/T | FTO | 0.24 | 1.04(1.02–1.06) | 7.5×10−4 | 0.18 | 1.04(0.98–1.11) | 0.23 | 1.04 (0.96–1.13) | 0.36 | 1.05 (0.95–1.16) | 0.37 | 0.81 |
| rs13329835 | 16 | 80650805 | G/A | CDYL2 | 0.22 | 1.08(1.05–1.10) | 2.1×10−16 | 0.62 | 1.04(0.98–1.09) | 0.20 | 1.07 (1.00–1.15) | 0.064 | 1.01 (0.93–1.10) | 0.74 | 0.087 |
| rs6504950 | 17 | 53056471 | G/A | COX11 | 0.73 | 1.06(1.04–1.09) | 2.3×10−13 | 0.64 | 1.03(0.98–1.09) | 0.23 | 1.04 (0.97–1.11) | 0.29 | 1.00 (0.92–1.08) | 0.95 | 0.45 |
| rs527616 | 18 | 24337424 | C/G | NA | 0.62 | 1.05(1.03–1.08) | 1.6×10−10 | 0.85 | 1.01(0.94–1.09) | 0.78 | 1.01 (0.91–1.11) | 0.86 | 0.95 (0.85–1.07) | 0.43 | 0.092 |
| rs1436904 | 18 | 24570667 | A/C | CHST9 | 0.60 | 1.04(1.02–1.06) | 3.2×10−8 | 0.74 | 1.00(0.94–1.06) | 0.97 | 1.00 (0.93–1.08) | 0.94 | 0.97 (0.89–1.06) | 0.53 | 0.71 |
| rs67397200e | 19 | 17401404 | G/C | - | 0.3 | 1.03(1.01–1.05) | 2.2×10−3 | 0.26 | 1.13(1.06–1.19) | 3.1×10−5 | 1.04 (0.97–1.12) | 0.29 | 1.19 (1.09–1.30) | 6.9×10−5 | 0.23 |
| rs4808801 | 19 | 18571141 | A/G | SSBP4:ISYNA1:ELL | 0.65 | 1.08(1.05–1.10) | 4.6×10−15 | 0.33 | 1.01(0.96–1.07) | 0.66 | 1.00 (0.93–1.07) | 0.94 | 0.99 (0.91–1.08) | 0.82 | 0.17 |
| rs3760982 | 19 | 44286513 | A/G | C19orf61:KCNN4: LYPD5:ZNF283 | 0.46 | 1.06(1.04–1.08) | 2.1×10−10 | 0.47 | 1.04(0.99–1.09) | 0.15 | 1.05 (0.98–1.12) | 0.18 | 0.97 (0.90–1.05) | 0.49 | 0.47 |
| rs2284378 | 20 | 32588095 | T/C | RALY | 0.31 | 1.08(1.05–1.12) | 1.3×10−6 | 0.16 | 1.00(0.94–1.08) | 0.89 | 0.97 (0.89–1.06) | 0.48 | 1.06 (0.95–1.17) | 0.31 | 0.42 |
| rs2823093 | 21 | 16520832 | G/A | NRIP1 | 0.74 | 1.09(1.06–1.11) | 6.8×10−16 | 0.57 | 1.01(0.97–1.07) | 0.56 | 1.00 (0.94–1.07) | 0.90 | 0.98 (0.90–1.06) | 0.57 | 0.47 |
| rs132390 | 22 | 29621477 | G/A | EMID1:RHBDD3: EWSR1 | 0.036 | 1.12(1.07–1.18) | 3.1×10−9 | 0.05 | 0.89(0.80–1.00) | 0.042 | 0.87 (0.75–1.01) | 0.067 | 0.90 (0.76–1.07) | 0.25 | 0.85 |
| rs6001930 | 22 | 40876234 | G/A | MKL1 | 0.11 | 1.12(1.09–1.16) | 8.8×10−19 | 0.13 | 1.05(0.97–1.13) | 0.21 | 1.09 (0.99–1.20) | 0.091 | 1.07 (0.95–1.19) | 0.27 | 0.24 |
SNP positions are based on GRCh37.
Risk/Reference Allele. Risk Allele is the allele associated with increased breast cancer risk in previous GWAS.
RAF, Risk Allele Frequency in controls of previous GWAS studies or in controls of AABC.
Heterogeneity between AABC, ROOT and AMBER.
OR reported for ER− breast cancer.
OR reported in African Americans.
OR reported in Asians.
To identify markers at known risk regions that might better define the index signals or serve as secondary, independent signals, fine-mapping analysis was conducted at each of the 70 regions (excluding 5p15 and 20q11, see Materials and Methods). Using region-specific thresholds, we observed associations of 7 markers with overall breast cancer or breast cancer subtypes at 6 regions (3p24, 12p11, 14q13, 16q12/FTO, 16q23, 19p13), while in two regions (11q13 and 16q12) we observed suggestive evidence of signals independent of the reported index variant (Supplementary Table S5). These regions are discussed below.
At 3p24, the index variant, rs4973768, was more strongly associated with ER− than ER+ disease in the initial GWAS (ER+: OR = 1.06, ER−: OR = 1.12, Phet = 0.022). Variant rs2370946, located in the intron of the NEK10 gene, with enhancer histone marks in ER+ breast cancer cells (i.e. HCC1954) and 155kb from the index variant, rs4973768, was found in association with ER+ breast cancer in women of African ancestry (ER+: OR = 1.17, P = 7.8×10−4; ER−: OR = 1.11, Phet = 0.058) (Supplementary Figure S1 & S2). Variant rs2370946 is correlated with the index in European populations, but not in African populations (EUR: r2 = 0.66; AFR: r2 = 0.01).
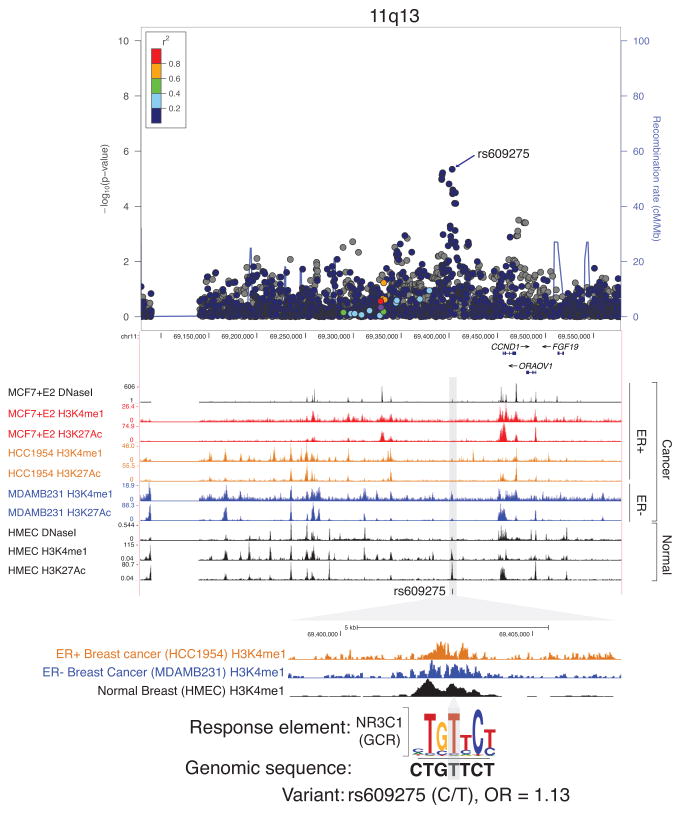
At 11q13, the same variant reported by Chen et al (rs609275: OR = 1.20, P = 1.0 × 10−5) (14) was identified as an independent secondary signal in this region (Supplementary Table S5). This variant was statistically significantly associated with overall breast cancer in women of African ancestry (OR = 1.13, P = 4.5 × 10−6; r2 with the index variant: 0.022 (EUR), 0.003 (AFR)) (Figure 1, Supplementary Figure S1). The variant rs609275, which resides in a gene desert region at 11q13, is located in a breast-specific active enhancer found not only in normal breast cells but also breast cancer cells (both ER+ and ER−). We observed that the motif of NR3C1 (aka GR, glucocorticoid receptor) is disrupted by the SNP; NR3C1 is known to inhibit MAPK activation by inducing MAPK1, possibly influencing breast cancer cell survival (45) (Figure 1; Supplementary Figure S2).
Figure 1. Regional plot and genome browser view of 11q13.
The chromosomal position (based on GRCh37) of SNPs on 11q13 against −log10 P-values for overall breast cancer is shown on the top plot. The blue arrow denotes the secondary signal rs609275. The purple circle denotes the index variant rs614367. SNPs surrounding the index variant are colored to indicate the LD structure using pairwise r2 in reference to rs614367 from the May 2012 EUR panel of 1000 Genomes. The plots were generated using LocusZoom (55). Genome browser views with epigenetic chromatin features in breast cells (MCF7, HCC1954, MDAMB231, HMEC) on 11q13 are generated using the UCSC genome browser (56). Below is a magnified view of rs609275 with selected enhancer chromatin marks and DNA sequence of a response element. The gray shading indicates the location of the variant rs609275.
At 12p11, the index variant, rs10771399, was statistically significantly associated with both ER+ and ER− breast cancer in the initial GWAS (46). No significant association was observed with overall breast cancer or breast cancer subtypes in women of African ancestry. Fine-mapping of this region in our African ancestry sample revealed two new variants, rs73094066 and rs805510, associated with overall and ER+ breast cancer, respectively (rs73094066 for overall breast cancer: OR = 1.11, P = 0.0027; rs805510 for ER+ disease: OR = 1.11, P = 0.0026). Both rs73094066 and rs805510 are correlated with the index variant (rs10771399) in European populations, but not in African populations (rs73094066: EUR r2 = 0.447 AFR r2 = 0.099; rs805510 EUR r2 = 0.912, AFR r2 = 0.005) (Supplementary Figure S1). A recent fine-mapping study on 12p11 detected a better marker, rs7297051, in Europeans (11). The better markers discovered in our study were modestly correlated with rs7297051 (rs73094066: EUR r2 = 0.084, AFR r2 = 0.003; rs805510 EUR r2 = 0.303, AFR r2 = 0.004). The variants rs73094066 and rs805510 are near enhancer histone marks, both found in breast cancer and normal breast cells, in the 12p11.22 gene desert region (Supplementary Figure S2).
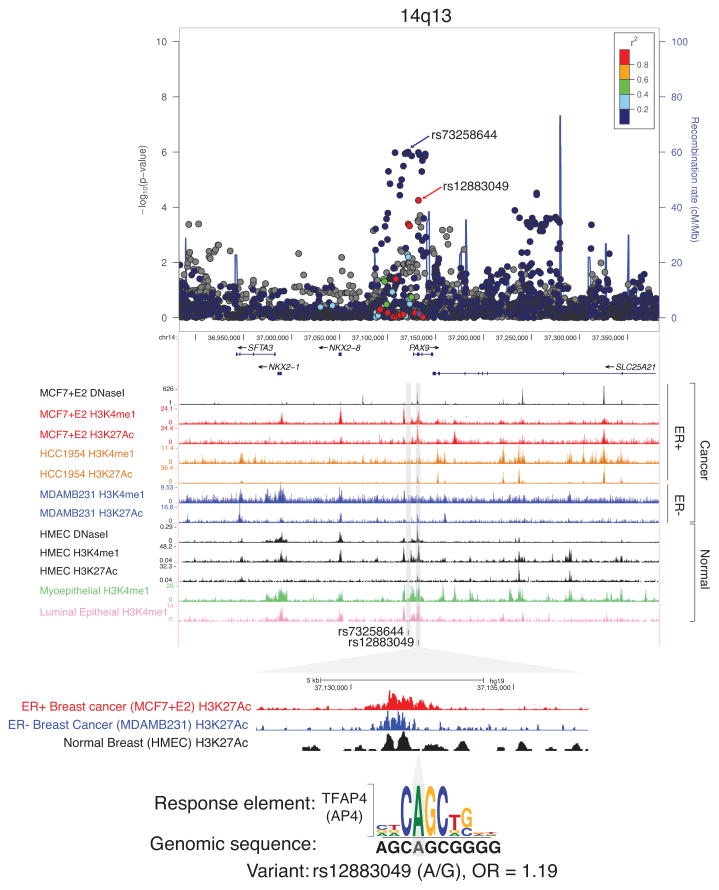
At 14q13, the index variant, rs2236007, was reported to be more strongly associated with ER+ than ER− breast cancer in the initial GWAS (ER+: OR = 1.10, P =1.9 × 10−10; ER−: OR = 1.04, P = 0.081, Phet = 0.015; Supplementary Table S4) (7). No association with the index variant could be detected in women of African ancestry (ER+: OR = 0.98, P = 0.72; ER−: OR = 0.94, P = 0.39). Through fine-mapping, the association with the most statistically significant P-value was observed with rs73258644 and ER+ disease (ER+: OR = 1.43, P = 1.0 × 10−6; ER−: OR = 1.02, P = 0.82). rs73258644 is a perfect proxy for rs17104923, which we previously reported in AABC as a potential independent signal (r2 = 1 in EUR and AFR), and shows no correlation with the index variant rs2236007 (EUR r2 = 0.008; AFR r2 = 0.002). Among markers correlated with the index variant, the strongest association was observed with rs12883049 and ER+ disease (OR = 1.19, P = 5.6 × 10−5) (Supplementary Figure S1). This variant, rs12883049, is located in the intron of PAX9 gene with enhancer histone marks and open chromatin marks in all breast cell lines, suggesting an important role of this variant (Figure 2, Supplementary Figure S2). We also found that the motif of TFAP4 (aka AP4) is disrupted by the SNP. AP4 is involved in the cell cycle and also activates cell migration and epithelial mesenchymal transition in breast cancer (47, 48). This variant is well correlated with the index variant in Europeans (r2 = 0.82), but not in women of African ancestry (r2 = 0.01). Variants rs73258644 and rs12883049 are modestly correlated (r2 = 0.35) and only rs73258644 remains statistically significant in conditional analyses with rs12883049 (P = 8.8 × 10−4) which suggests that rs73258644 is the best marker in the region relevant to women of African ancestry (Supplementary Table S6).
Figure 2. Regional plot and genome browser view of 14q13.
The chromosomal position (based on GRCh37) of SNPs on 14q13 against −log10 P-values for ER+ breast cancer is shown. The blue arrow denotes the signal rs73258644 and the red arrow denotes rs12883049, which is a better marker of the index signal. The purple circle denotes the index variant rs2236007. SNPs surrounding the index variant are colored to indicate the LD structure using pairwise r2 in reference to rs2236007 from the May 2012 EUR panel of 1000 Genomes. The plots were generated using LocusZoom (55). Genome browser views with epigenetic chromatin features in breast cells (MCF7, HCC1954, MDAMB231, HMEC) on 14q13 are generated using the UCSC genome browser (56). Below is a magnified view of rs73258644 with selected enhancer chromatin marks and DNA sequence of a response element. The gray shading indicates the location of the variant rs12883049.
At 16q12/TOX3, the index variant rs3803662 was identified initially in association with ER+ disease (12). This variant was not associated with breast cancer subtypes in women of African ancestry (Supplementary Table S4). Our fine-mapping analysis of this region revealed a risk variant in the intron of TOX3 gene, rs35850695 (r2 = 0.89 in EUR), that was more strongly associated with ER+ breast cancer (ER+: OR = 1.25, P = 2.4 × 10−5; ER−: OR = 1.07, P = 0.33; Phet = 0.033; Supplementary Table S5). However, the most statistically significantly associated risk variant in this region was rs3104791, which is located in the intron of long noncoding RNA (lncRNA), LINC00918 (OR = 1.18 for ER+ disease, P = 1.8 × 10−6) (Supplementary Figure S2). This variant is moderately correlated with the index (rs3803662) in both women of European and African ancestry (EUR: r2 = 0.28; AFR r2 = 0.20) and is also moderately correlated with rs35850695 in Europeans, but not in women of African ancestry (EUR: r2 = 0.24; AFR r2 = 0.018). A second potentially independent signal, rs3112565, was also noted (OR = 1.19, P = 2.3 × 10−5), which is a perfect proxy (r2 = 1 in AFR) for rs3112572 (14) and rs3104746 reported previously (49) (Supplementary Table S6). In conditional analyses of these three signals, rs35850695 (P = 5.2 × 10−5) and rs3112565 (P = 0.0011) remained as independent signals for ER+ disease, but not rs3104791 (P = 0.054) (Supplementary Table S6). Haplotypes containing the risk variant for rs3104791 were statistically significantly associated with risk together with either the risk alleles of rs3112565 and/or rs35850695, but not alone (OR = 1.03; P = 0.54; Supplementary Table S6). The variant rs35850695 is located in the intron of TOX3 gene, whereas the variants rs3112565 and rs310479 are located in the intron of LINC00918. The variant rs3112565 is also found in ER+ cancer specific enhancer regions, annotated by histone marks, H3K4me1 and H3K27Ac (Supplementary Figure S2).
At 16q12/FTO, two independent signals (rs17817449 and rs11075995) were discovered to be associated with breast cancer risk in previous GWAS and rs11075995 was identified as an ER− specific variant (7). In women of African ancestry, rs17817449 showed a statistically significant association with both overall breast cancer and ER+ disease (overall: OR = 1.07, P = 0.012; ER+: OR = 1.08, P = 0.029). We observed an association with rs62048370, that was statistically significantly and more strongly associated with ER+ breast cancer (overall: OR = 1.29, P = 0.00032; ER+: OR = 1.59, P = 3.0 × 10−6; ER−: OR = 1.04, P = 0.72). Variant rs62048370 is not correlated with either of the index variants in European or African populations (rs17817449: EUR r2 < 0.001, AFR r2 = 0.004; rs11075995: EUR r2 < 0.001, AFR r2 = 0.007) (Supplementary Figure S1 and Supplementary Table S5). This variant also overlaps with enhancer histone marks in ER+ breast cancer and normal breast cell lines, which are in close proximity to open chromatin regions in which transcription factors such as FOXA1, GATA3, and ESR1 bind (Supplementary Figure S2).
At 16q23, the index variant rs13329835 was reported to be more strongly associated with ER+ disease in the initial GWAS (7). Through fine-mapping, we identified another variant, rs9940301, that is highly correlated with the index variant in Europeans (r2 = 0.84), and was statistically significantly associated with ER+ breast cancer in women of African ancestry (OR = 1.13, P = 8.5 × 10−4; Supplementary Table S5 and Supplementary Figure S1). The variant rs9940301 is in the intron of the CDYL2 gene, and encodes a chromodomain protein, which interacts with histone H3K9me3 (Supplementary Figure S2) (50).
At the ER− risk region 19p13, variant rs11668840, which is correlated with the index SNP in Europeans (rs67397200; r2 = 0.49), was the most statistically significantly associated marker for ER− breast cancer (OR = 1.25, P = 3.1 × 10−8; Supplementary Figure S1, Supplementary Table S4 and S5). The variant rs11668840 is 1.2kb downstream of the transcription termination site of the ANKLE1 gene and is not located within any regions of open chromatin (Supplementary Figure S2).
Discussion
The majority of GWAS-identified risk variants for breast cancer are common in women of African ancestry with directions of effect that are consistent with the discovery populations. However, in this sample, which is the largest breast cancer genetics study ever conducted in women of African ancestry (6,522 cases and 7,643 controls), only 12 variants were directionally consistent with previous GWAS and nominally statistically significant at P < 0.05. In fine-mapping, we were successful in identifying seven markers for overall breast cancer or breast cancer subtype in six regions (3p24, 12p11, 14q13, 16q12/FTO, 16q23, 19p13) that were more likely (than the index variant) to capture the breast cancer association in this population. In another two regions (11q13 and 16q12/TOX3) we identified risk variants independent of the index signal. Among these regions harboring better markers or independent signals, only at 19p13, was the index variant also significantly associated with breast cancer risk.
The 74 variants analyzed in this study were reported to have an average odds ratio of 1.09, with only 17 (23%) having ORs >1.10. Of the 61 SNPs that were not statistically significant in African Americans, statistical power to detect the previously reported effect sizes for overall breast cancer was ≥ 80% for 29 SNPs (48%). While reasonable statistical power was noted for roughly 50% of these regions, the inability to achieve statistical significance for the majority of these loci is likely due to differences in LD structure between populations of European and African ancestry. Statistical power in fine-mapping analyses is even more severely limited as we employed conservative locus-specific alpha levels to limit the number of false-positive associations. Statistical power to detect associations as large as those of the index signals while adjusting for multiple comparisons in the fine mapping was ≥ 80% at only 13 of the 70 regions (Supplementary Table S2). It is important to note that the markers we highlighted in each region only indicate whether the region replicates in African ancestry populations. There is a high degree of variability in the association statistics (ORs, P values and standard errors) due to many factors including genotyping success rate and imputation quality, which has an impact on the ranking of associated correlated SNPs.
To further prioritize variants for functional follow-up testing, we mapped the most strongly associated variants relative to epigenomic datasets (see Materials and Methods). For the better markers or independent signals we identified in this study, 7 overlapped with enhancer histone marks (Supplementary Table S3). Additionally, we discovered that some of the better marker/independent signals specific in ER+ breast cancer subtypes were found in ER+ breast cancer specific enhancers (e.g. rs2370946 at 3p24). On the other hand, some of the better markers or independent signals associated with overall breast cancer risk (both ER+ and ER−) were found in putative breast enhancers common in both ER+ and ER− breast cancer cells (e.g. rs609275 in 11q13). The underlying risk variants may play different roles and have unique mechanisms to increase breast cancer risk, however we may deduce that subtype specific enhancer activity might be tightly linked with some of these risk regions.
The most statistically significant associations in women of African ancestry identified in both previous studies as well as the current investigation were with variants on 11q13, 14q13, 16q12/TOX3 and 19p13 (14, 15). At 11q13, the putative novel signal locates 53kb upstream of CCND1 (Cyclin D1). Cyclin D1 plays a key role in cell cycle regulation and is one of the most commonly overexpressed proteins in breast tumors (51). At 14q13, variants were located in the gene PAX9 (paired box 9), which has been shown to be required for the growth and survival of breast cancer cells (52). At 16q12, the signals are located within the intron of a lncRNA, LINC00918, and the TOX3 (TOX high mobility group box family member 3) gene, which may be involved in the bending and unwinding of DNA and altering chromatin structure (53). At 19p13, the risk variant is located near the genes BABAM1 (BRISC and BRCA1-A complex member 1), ANKLE1 (Ankyrin Repeat And LEM Domain-Containing Protein 1) and ABHD8 (abhydrolase domain containing 8). BABAM1 is the best candidate that may be influenced by genetic variation in the region given its interaction with BRCA1 (54).
In conclusion, 54 (73%) of the 74 breast cancer risk variants examined in women of African ancestry had effects that were directionally consistent with those previously reported, with 12 being nominally statistically significant. These findings support prior studies indicating that the majority of established breast cancer risk loci found in populations of European and Asian ancestry are also likely to be susceptibility regions for women of African ancestry. In six regions we observed suggestive evidence of common alleles that may better characterize the association with breast cancer in women of African ancestry. Despite the sample size of the current effort, which includes all existing genetic studies of breast cancer in women of African ancestry globally, substantially larger studies, including multiethnic studies, will be needed to fully understand the genetic architecture of breast cancer in women of African ancestry.
Supplementary Material
Acknowledgments
This research was funded by the National Institutes of Health (NIH) and Foundation grants: P01 CA151135, R01 CA058420, UM1 CA164974, R01 CA098663, R01 CA100598, R01 CA185623, UM1 CA164973, R01 CA54281, R01 CA063464, R01 CA190182, P50 CA58223, U01 CA179715, R01 CA142996, P50 CA125183, and R01 CA89085, the Department of Defense Breast Cancer Research Program, Era of Hope Scholar Award Program W81XWH-08-1-0383 (AABC); the Susan G. Komen for the Cure Foundation; the Breast Cancer Research Foundation; and the University Cancer Research Fund of North Carolina. Pathology data were obtained from numerous state cancer registries (Arizona, California, Colorado, Connecticut, Delaware, District of Columbia, Florida, Georgia, Hawaii, Illinois, Indiana, Kentucky, Louisiana, Maryland, Massachusetts, Michigan, New Jersey, New York, North Carolina, Oklahoma, Pennsylvania, South Carolina, Tennessee, Texas, Virginia). Studies in AABC were supported by National Institute for Child Health and Development contract NO1-HD-3-3175 (CARE), NIH grant CA100374 and the Biospecimen Core Lab that is supported in part by the Vanderbilt-Ingram Cancer Center (CA68485) (NBHS), by NIH grant CA73629 (WFBC), NIH grant CA77305 and United States Army Medical Research Program grant DAMD17-96-6071 (SFBCS), by NIH grant CA164920 (NC-BCFR). The Breast Cancer Family Registry (BCFR) was supported by grant UM1 CA164920 from the USA National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. Genotyping of the PLCO samples was funded by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, NCI, NIH.
References
- 1.Dunning AM, Michailidou K, Kuchenbaecker KB, Thompson D, French JD, Beesley J, et al. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat Genet. 2016;48:374–86. doi: 10.1038/ng.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–8. 8e1–2. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glubb DM, Maranian MJ, Michailidou K, Pooley KA, Meyer KB, Kar S, et al. Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1. Am J Hum Genet. 2015;96:5–20. doi: 10.1016/j.ajhg.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrenson K, Kar S, McCue K, Kuchenbaeker K, Michailidou K, Tyrer J, et al. Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast-ovarian cancer susceptibility locus. Nat Commun. 2016;7:12675. doi: 10.1038/ncomms12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long J, Cai Q, Sung H, Shi J, Zhang B, Choi JY, et al. Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet. 2012;8:e1002532. doi: 10.1371/journal.pgen.1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47:373–80. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–61. 61e1–2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orr N, Dudbridge F, Dryden N, Maguire S, Novo D, Perrakis E, et al. Fine-mapping identifies two additional breast cancer susceptibility loci at 9q31.2. Hum Mol Genet. 2015;24:2966–84. doi: 10.1093/hmg/ddv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi J, Sung H, Zhang B, Lu W, Choi JY, Xiang YB, et al. New breast cancer risk variant discovered at 10q25 in East Asian women. Cancer Epidemiol Biomarkers Prev. 2013;22:1297–303. doi: 10.1158/1055-9965.EPI-12-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyszynski A, Hong CC, Lam K, Michailidou K, Lytle C, Yao S, et al. An intergenic risk locus containing an enhancer deletion in 2q35 modulates breast cancer risk by deregulating IGFBP5 expression. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng C, Guo X, Long J, Kuchenbaecker KB, Droit A, Michailidou K, et al. Identification of independent association signals and putative functional variants for breast cancer risk through fine-scale mapping of the 12p11 locus. Breast Cancer Res. 2016;18:64. doi: 10.1186/s13058-016-0718-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 14.Chen F, Chen GK, Millikan RC, John EM, Ambrosone CB, Bernstein L, et al. Fine-mapping of breast cancer susceptibility loci characterizes genetic risk in African Americans. Hum Mol Genet. 2011;20:4491–503. doi: 10.1093/hmg/ddr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y, Stram DO, Rhie SK, Millikan RC, Ambrosone CB, John EM, et al. A comprehensive examination of breast cancer risk loci in African American women. Hum Mol Genet. 2014;23:5518–26. doi: 10.1093/hmg/ddu252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen F, Chen GK, Stram DO, Millikan RC, Ambrosone CB, John EM, et al. A genome-wide association study of breast cancer in women of African ancestry. Hum Genet. 2013;132:39–48. doi: 10.1007/s00439-012-1214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer JR, Viscidi E, Troester MA, Hong CC, Schedin P, Bethea TN, et al. Parity, lactation, and breast cancer subtypes in African American women: results from the AMBER Consortium. J Natl Cancer Inst. 2014:106. doi: 10.1093/jnci/dju237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer JR, Ambrosone CB, Olshan AF. A collaborative study of the etiology of breast cancer subtypes in African American women: the AMBER consortium. Cancer Causes Control. 2014;25:309–19. doi: 10.1007/s10552-013-0332-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y, Ogundiran TO, Falusi AG, Nathanson KL, John EM, Hennis AJ, et al. Fine mapping of breast cancer genome-wide association studies loci in women of African ancestry identifies novel susceptibility markers. Carcinogenesis. 2013;34:1520–8. doi: 10.1093/carcin/bgt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenberg L, Adams-Campbell L, Palmer JR. The Black Women’s Health Study: a follow-up study for causes and preventions of illness. J Am Med Womens Assoc. 1995;50:56–8. [PubMed] [Google Scholar]
- 21.Ambrosone CB, Ciupak GL, Bandera EV, Jandorf L, Bovbjerg DH, Zirpoli G, et al. Conducting Molecular Epidemiological Research in the Age of HIPAA: A Multi-Institutional Case-Control Study of Breast Cancer in African-American and European-American Women. J Oncol. 2009;2009:871250. doi: 10.1155/2009/871250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman B, Moorman PG, Millikan R, Qaqish BF, Geradts J, Aldrich TE, et al. The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast Cancer Res Treat. 1995;35:51–60. doi: 10.1007/BF00694745. [DOI] [PubMed] [Google Scholar]
- 23.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 26.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet. 2011;43:1210–4. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddiq A, Couch FJ, Chen GK, Lindstrom S, Eccles D, Millikan RC, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5373–84. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhie SK, Hazelett DJ, Coetzee SG, Yan C, Noushmehr H, Coetzee GA. Nucleosome positioning and histone modifications define relationships between regulatory elements and nearby gene expression in breast epithelial cells. BMC Genomics. 2014;15:331. doi: 10.1186/1471-2164-15-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhie SK, Coetzee SG, Noushmehr H, Yan C, Kim JM, Haiman CA, et al. Comprehensive functional annotation of seventy-one breast cancer risk Loci. PLoS One. 2013;8:e63925. doi: 10.1371/journal.pone.0063925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhie SK, Guo Y, Tak YG, Yao L, Shen H, Coetzee GA, et al. Identification of activated enhancers and linked transcription factors in breast, prostate, and kidney tumors by tracing enhancer networks using epigenetic traits. Epigenetics Chromatin. 2016;9:50. doi: 10.1186/s13072-016-0102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gascard P, Bilenky M, Sigaroudinia M, Zhao J, Li L, Carles A, et al. Epigenetic and transcriptional determinants of the human breast. Nat Commun. 2015;6:6351. doi: 10.1038/ncomms7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blahnik KR, Dou L, O’Geen H, McPhillips T, Xu X, Cao AR, et al. Sole-Search: an integrated analysis program for peak detection and functional annotation using ChIP-seq data. Nucleic Acids Res. 2010;38:e13. doi: 10.1093/nar/gkp1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taberlay PC, Statham AL, Kelly TK, Clark SJ, Jones PA. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res. 2014;24:1421–32. doi: 10.1101/gr.163485.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coetzee SG, Coetzee GA, Hazelett DJ. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015;31:3847–9. doi: 10.1093/bioinformatics/btv470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–7. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Zhuang J, Iyer S, Lin XY, Greven MC, Kim BH, et al. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2013;41:D171–6. doi: 10.1093/nar/gks1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–92. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu W, Pew T, Zou M, Pang D, Conzen SD. Glucocorticoid receptor-induced MAPK phosphatase-1 (MPK-1) expression inhibits paclitaxel-associated MAPK activation and contributes to breast cancer cell survival. J Biol Chem. 2005;280:4117–24. doi: 10.1074/jbc.M411200200. [DOI] [PubMed] [Google Scholar]
- 46.Ghoussaini M, Fletcher O, Michailidou K, Turnbull C, Schmidt MK, Dicks E, et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat Genet. 2012;44:312–8. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S, Chiu SK. AP4 activates cell migration and EMT mediated by p53 in MDA-MB-231 breast carcinoma cells. Mol Cell Biochem. 2015;407:57–68. doi: 10.1007/s11010-015-2454-7. [DOI] [PubMed] [Google Scholar]
- 48.Jung P, Menssen A, Mayr D, Hermeking H. AP4 encodes a c-MYC-inducible repressor of p21. Proc Natl Acad Sci U S A. 2008;105:15046–51. doi: 10.1073/pnas.0801773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruiz-Narvaez EA, Rosenberg L, Cozier YC, Cupples LA, Adams-Campbell LL, Palmer JR. Polymorphisms in the TOX3/LOC643714 locus and risk of breast cancer in African-American women. Cancer Epidemiol Biomarkers Prev. 2010;19:1320–7. doi: 10.1158/1055-9965.EPI-09-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morvan D, Leroy-Willig A, Malgouyres A, Cuenod CA, Jehenson P, Syrota A. Simultaneous temperature and regional blood volume measurements in human muscle using an MRI fast diffusion technique. Magn Reson Med. 1993;29:371–7. doi: 10.1002/mrm.1910290313. [DOI] [PubMed] [Google Scholar]
- 51.He Y, Liu Z, Qiao C, Xu M, Yu J, Li G. Expression and significance of Wnt signaling components and their target genes in breast carcinoma. Mol Med Rep. 2014;9:137–43. doi: 10.3892/mmr.2013.1774. [DOI] [PubMed] [Google Scholar]
- 52.Muratovska A, Zhou C, He S, Goodyer P, Eccles MR. Paired-Box genes are frequently expressed in cancer and often required for cancer cell survival. Oncogene. 2003;22:7989–97. doi: 10.1038/sj.onc.1206766. [DOI] [PubMed] [Google Scholar]
- 53.O’Flaherty E, Kaye J. TOX defines a conserved subfamily of HMG-box proteins. BMC Genomics. 2003;4:13. doi: 10.1186/1471-2164-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–28. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.