Figure 1. Genomic organization of the IgH locus and immunoglobulin structure.
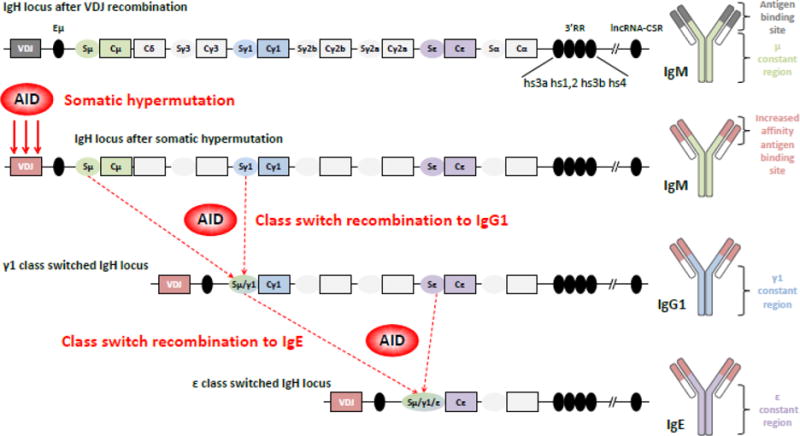
Top. The mouse IgH locus is represented after VDJ recombination (not to scale). The recombined VDJ gene and constant (C) genes are represented as outlined boxes. Horizontally-oriented ovals portray switch (S) regions (preceded by promoters and I exons, not shown) preceding each constant gene (excepted Cδ). Black ovals portray regulatory elements: intronic enhancer μ (Eμ), 3′ regulatory region (3′RR) and the newly identified lncRNA-CSR region, located approximately 2.6 mega-bases downstream of the 3′RR. The resulting immunoglobulin (Ig) protein is IgM, shown on the right.
Middle. After B cell activation, AID is expressed and induces its mutagenic activity on the VDJ gene, allowing production of IgM antibodies with increased affinity for the antigen. Then AID is targeted to S regions to initiate double strand breaks, thereby permitting recombination between two S regions, here Sμ and Sγ1. This process generates high affinity IgG1 antibodies and a diversified B cell repertoire.
Bottom. Cells can re-express AID after re-exposure to antigen and undergo a second class switch recombination event, from γ1 to ε here, to produce high affinity IgE antibodies (direct class switching from μ to ε is also possible). All these AID-mediated events shape the Ig repertoire to improve antigen affinity and to adapt the Ig class for an optimal immune response.
