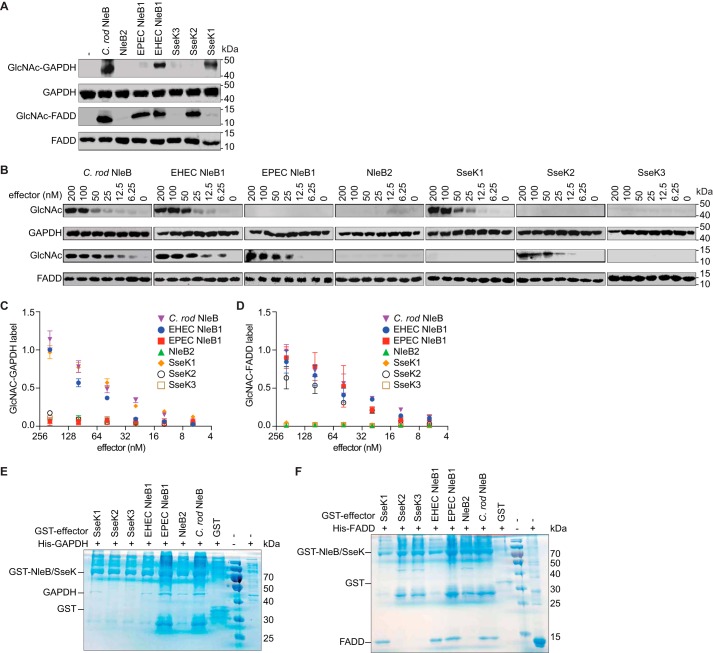
Figure 2.
NleB/SseK orthologs differentially glycosylate GAPDH and FADD. A, in vitro GlcNAcylation assays. NleB/SseK enzymes (200 nm) were incubated for 4 h at room temperature with either with GAPDH or FADD (1 μm) in 50 mm Tris-HCl, pH 7.4, 10 mm UDP-GlcNAc, 10 mm MnCl2, and 1 mm DTT. The samples were then subjected to Western blot detection using an anti-R-GlcNAc monoclonal antibody. B, titration of NleB/SseK orthologs in GAPDH and FADD GlcNAcylation assays. Serial dilutions of NleB/SseK enzymes (6.25–200 nm) were incubated with either GAPDH or FADD (1 μm) and then subjected to Western blot detection using an anti-R-GlcNAc monoclonal antibody. C, quantification of GAPDH glycosylation. The data shown in B were quantified by normalizing GlcNAc-GAPDH signals to total GAPDH signals. The data shown are representative of three independent experiments. D, quantification of FADD glycosylation. The data shown in B were quantified by normalizing GlcNAc-FADD signals to total FADD signals. The data shown are representative of three independent experiments. E and F, pulldown assays to detect binding between the NleB/SseK orthologs and GAPDH (E) or FADD (F). His-GAPDH or His-FADD were individually incubated with each NleB/SseK ortholog then subjected to GST pulldown assays using glutathione-Sepharose beads (GE Healthcare). Protein complexes were eluted with 10 mm reduced glutathione followed by SDS-PAGE analysis. GST was used as negative control. C. rod, C. rodentium.